|
|
|
|

|
| Sample: |
Alarmin release inhibitor (Δ1-62) monomer, 26 kDa Heligmosomoides polygyrus protein
Interleukin-33 (L179V) monomer, 18 kDa Mus musculus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin-33 (L179V) monomer, 18 kDa Mus musculus protein
Alarmin release inhibitor (Δ1-125; N175Q, N190Q) monomer, 19 kDa Heligmosomoides polygyrus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-homologous end-joining factor 1 dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-tris, 150 mM KCl, 1 mM EDTA, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Mar 13
|
Multivalent interactions of the disordered regions of XLF and XRCC4 foster robust cellular NHEJ and drive the formation of ligation-boosting condensates in vitro.
Nat Struct Mol Biol (2024)
Vu DD, Bonucci A, Brenière M, Cisneros-Aguirre M, Pelupessy P, Wang Z, Carlier L, Bouvignies G, Cortes P, Aggarwal AK, Blackledge M, Gueroui Z, Belle V, Stark JM, Modesti M, Ferrage F
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA repair protein XRCC4 dimer, 77 kDa Homo sapiens protein
DNA ligase 4 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-tris, 150 mM KCl, 1 mM EDTA, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Mar 13
|
Multivalent interactions of the disordered regions of XLF and XRCC4 foster robust cellular NHEJ and drive the formation of ligation-boosting condensates in vitro.
Nat Struct Mol Biol (2024)
Vu DD, Bonucci A, Brenière M, Cisneros-Aguirre M, Pelupessy P, Wang Z, Carlier L, Bouvignies G, Cortes P, Aggarwal AK, Blackledge M, Gueroui Z, Belle V, Stark JM, Modesti M, Ferrage F
|
| RgGuinier |
5.8 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
248 |
nm3 |
|
|
|
|
|
|
|
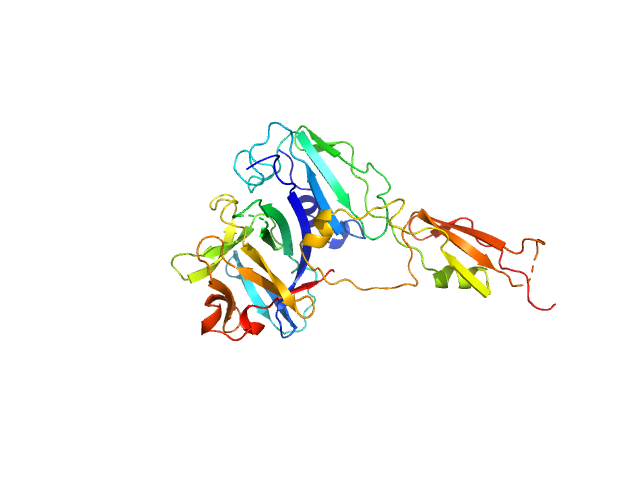
| Sample: |
DNA repair protein XRCC4 monomer, 15 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-tris, 150 mM KCl, 1 mM EDTA, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 20
|
Multivalent interactions of the disordered regions of XLF and XRCC4 foster robust cellular NHEJ and drive the formation of ligation-boosting condensates in vitro.
Nat Struct Mol Biol (2024)
Vu DD, Bonucci A, Brenière M, Cisneros-Aguirre M, Pelupessy P, Wang Z, Carlier L, Bouvignies G, Cortes P, Aggarwal AK, Blackledge M, Gueroui Z, Belle V, Stark JM, Modesti M, Ferrage F
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
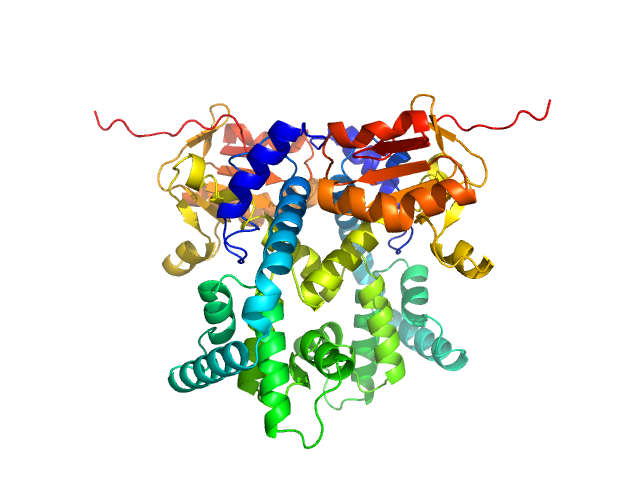
| Sample: |
CDAN1-interacting nuclease 1 dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2021 Sep 2
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
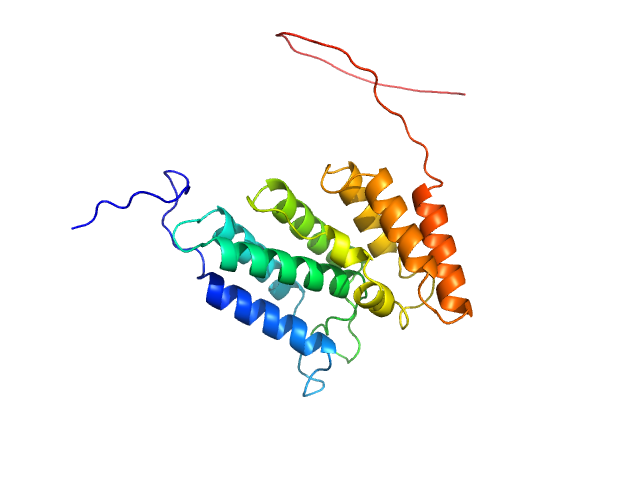
| Sample: |
Codanin-1 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Mar 5
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Codanin-1 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Mar 5
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
|
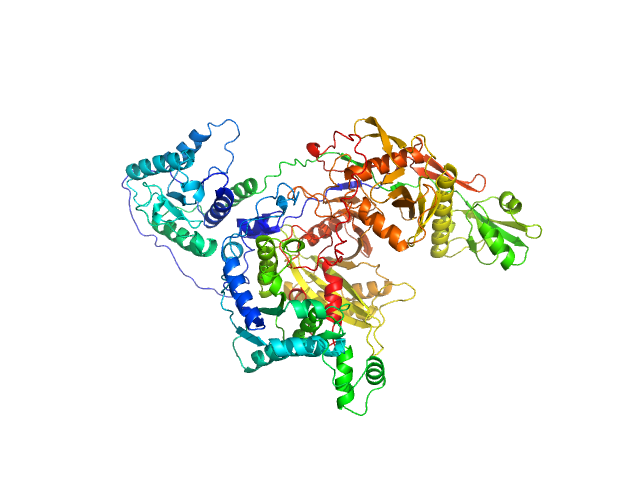
| Sample: |
TIR domain-containing protein monomer, 57 kDa Rhodopseudomonas palustris (strain … protein
Uncharacterized protein (PIWI) monomer, 56 kDa Rhodopseudomonas palustris (strain … protein
|
| Buffer: |
20 mM Tris-HCl, pH8.0, 200 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 27
|
Complex of Rhodopseudomonas palustris pAgo with TIR-like effector protein
Elena Manakova
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HbP1 trimer, 56 kDa Legionella pneumophila protein
|
| Buffer: |
20 mM Tris–HCl pH 8.0, 200 mM NaCl, 5 mM EDTA, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
The Legionella collagen-like protein employs a distinct binding mechanism for the recognition of host glycosaminoglycans.
Nat Commun 15(1):4912 (2024)
Rehman S, Antonovic AK, McIntire IE, Zheng H, Cleaver L, Baczynska M, Adams CO, Portlock T, Richardson K, Shaw R, Oregioni A, Mastroianni G, Whittaker SB, Kelly G, Lorenz CD, Fornili A, Cianciotto NP, Garnett JA
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
105 |
nm3 |
|
|