|
|
|
|
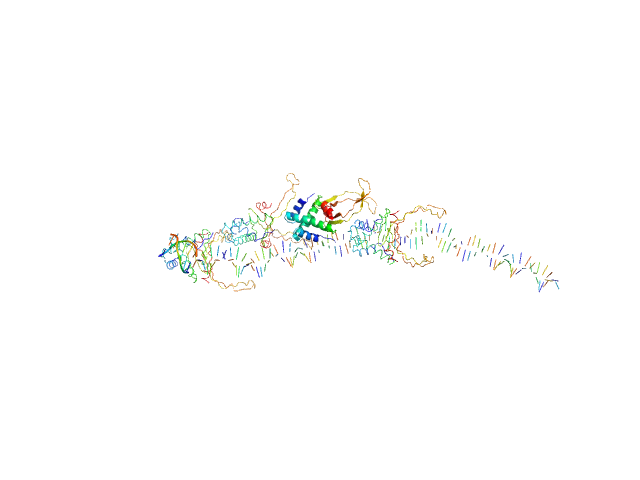
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E38K/V42L double mutant octamer, 76 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Apr 23
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling (supplementary)
Soumya G Remesh
|
| RgGuinier |
5.8 |
nm |
| Dmax |
28.1 |
nm |
| VolumePorod |
296 |
nm3 |
|
|
|
|
|
|
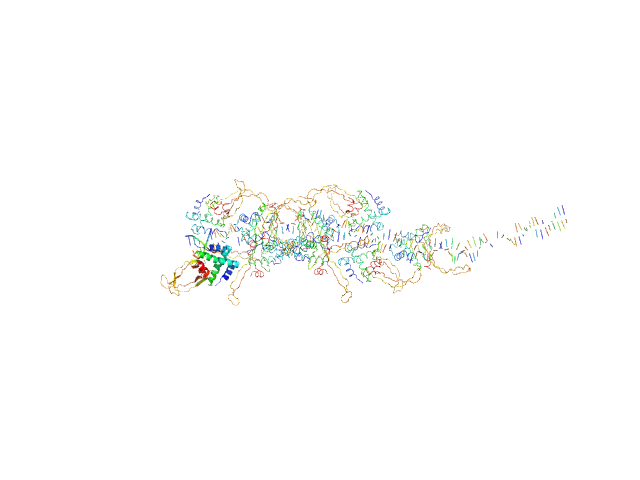
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E38K/V42L double mutant 16-mer, 153 kDa Linked to wild-type … protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Apr 23
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling (supplementary)
Soumya G Remesh
|
| RgGuinier |
6.3 |
nm |
| Dmax |
27.3 |
nm |
| VolumePorod |
401 |
nm3 |
|
|
|
|
|
|
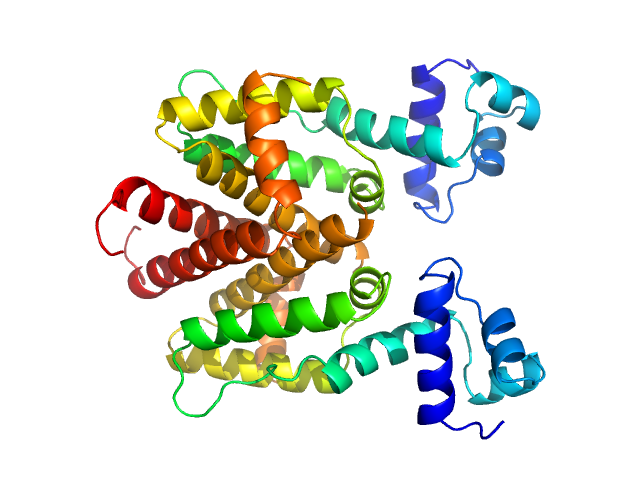
|
| Sample: |
Tetracycline repressor (class D) dimer, 47 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris/HCl 150 mM NaCl 10 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom :140404 (2020)
Palm GJ, Buchholz I, Werten S, Girbardt B, Berndt L, Delcea M, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
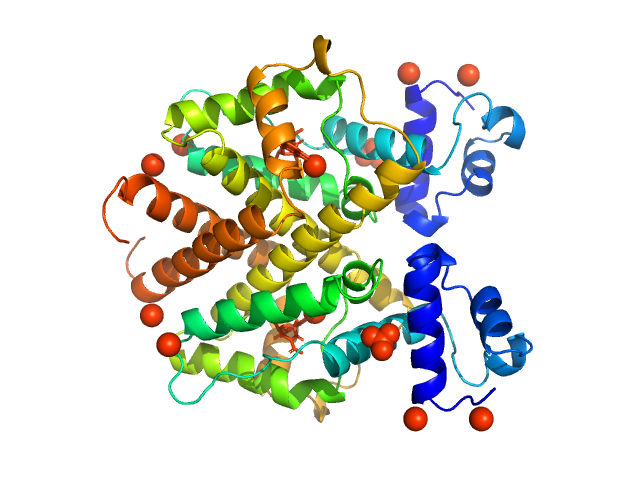
|
| Sample: |
Tetracycline repressor (class D) dimer, 47 kDa Escherichia coli protein
5a,6-anhydrotetracycline dimer, 1 kDa
|
| Buffer: |
50 mM Tris/HCl 150 mM NaCl 10 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom :140404 (2020)
Palm GJ, Buchholz I, Werten S, Girbardt B, Berndt L, Delcea M, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
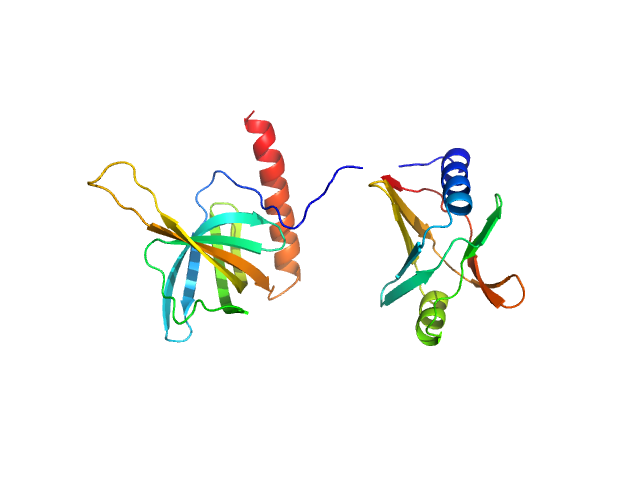
| Sample: |
Flagellar brake protein YcgR monomer, 29 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150mM NaCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jan 4
|
Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility in Escherichia coli.
J Biol Chem 295(3):808-821 (2020)
Hou YJ, Yang WS, Hong Y, Zhang Y, Wang DC, Li DF
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
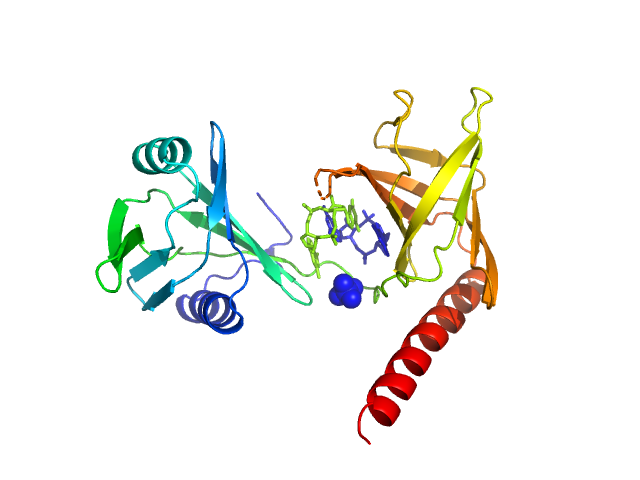
| Sample: |
Flagellar brake protein YcgR in complex with c-di-GMP monomer, 29 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150mM NaCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jan 4
|
Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility in Escherichia coli.
J Biol Chem 295(3):808-821 (2020)
Hou YJ, Yang WS, Hong Y, Zhang Y, Wang DC, Li DF
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 100 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Oct 28
|
Polymorphic Protective Dps-DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering.
Biomolecules 10(1) (2019)
Kamyshinsky R, Chesnokov Y, Dadinova L, Mozhaev A, Orlov I, Petoukhov M, Orekhov A, Shtykova E, Vasiliev A
|
|
|
|
|
|
|
|
| Sample: |
Lipid A export ATP-binding/permease protein MsbA - Nucleotide binding domain monomer, 27 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5 mM MgCl2, 0.45 mM Mg2+-ATP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 8
|
Structural Kinetics of MsbA Investigated by Stopped-Flow Time-Resolved Small-Angle X-Ray Scattering.
Structure (2019)
Josts I, Gao Y, Monteiro DCF, Niebling S, Nitsche J, Veith K, Gräwert TW, Blanchet CE, Schroer MA, Huse N, Pearson AR, Svergun DI, Tidow H
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
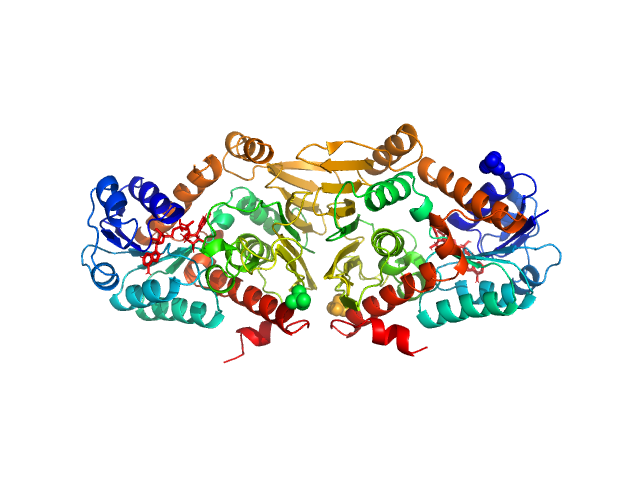
| Sample: |
Endoribonuclease E tetramer, 247 kDa Escherichia coli protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
468 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Escherichia coli YjhC dimer, 86 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, 5 % (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 12
|
On the structure and function of Escherichia coli YjhC: an oxidoreductase involved in bacterial sialic acid metabolism.
Proteins (2019)
Horne CR, Kind L, Davies JS, Dobson RCJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
130 |
nm3 |
|
|