|
|
|
|
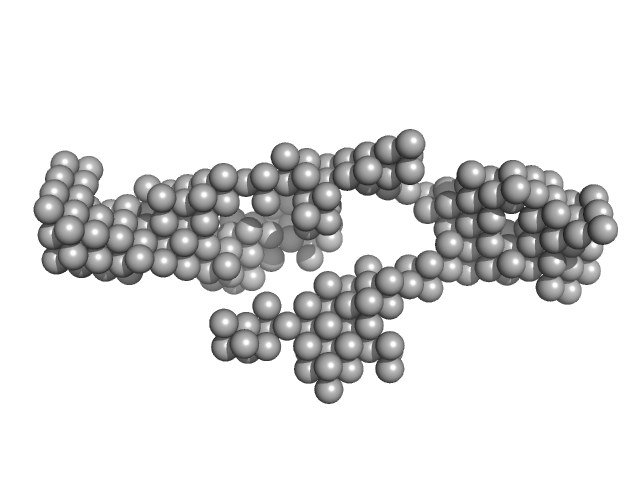
|
| Sample: |
7SL Signal Reocgnition Particle RNA monomer, 98 kDa Homo sapiens RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 15 mM KCl, 5% Glycerol, 3 mM MgCl2, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 21
|
Solution Structure Determination and Biophysical Studies of 7SK RNP and 7SL SRP RNAs.
RNA (2025)
D'Souza MH, Pereira HS, Tersteeg S, Vasudeva G, Siddiqui MQ, Agunlejika T, Kerr L, Girodat D, Patel T
|
| RgGuinier |
6.7 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
300 |
nm3 |
|
|
|
|
|
|
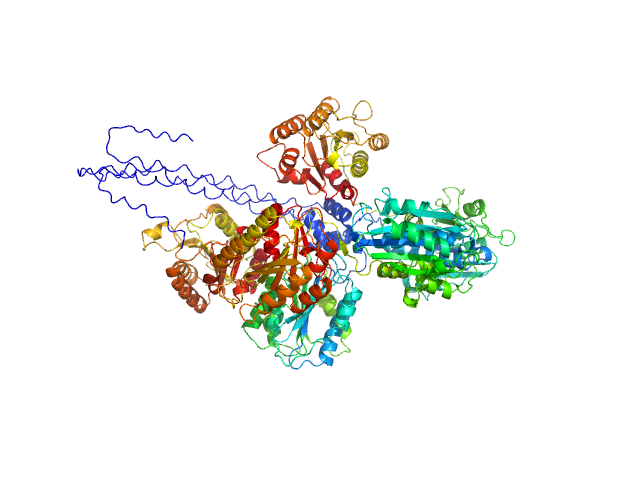
|
| Sample: |
Collagen alpha-1(VI) chain, 52 kDa Homo sapiens protein
Collagen alpha-2(VI) chain (R680H, K966N, Q967E), 53 kDa Homo sapiens protein
Collagen alpha-3(VI) chain (D2357R, K2367R, D2431V, R2441T, R2609A, R2610A), 54 kDa Homo sapiens protein
|
| Buffer: |
Tris-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Jul 27
|
Collagen VI Microfibril Structure Reveals the Mechanism for Heterotrimerisation and the Clustering of Inherited Pathogenic Mutations
Mark H. Becker
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.7 |
nm |
| VolumePorod |
478 |
nm3 |
|
|
|
|
|
|
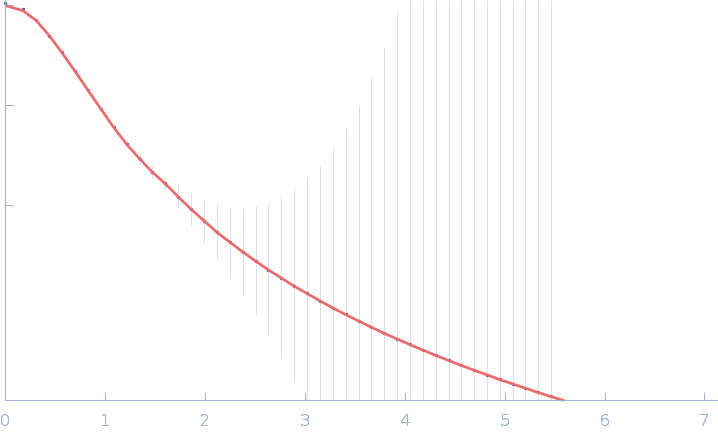
|
| Sample: |
Fas-activated serine/threonine kinase (amino acids 76-549) monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5% (v/v) Glycerol, 200 mM Ammonium Sulphate, 50 mM NaCl, 0.5 mM TCEP, 10 mM Glutamic acid, 10 mM Arginine;, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 31
|
Human FASTK preferentially binds single-stranded and G-rich RNA.
FEBS J (2025)
Dawidziak DM, Dzadz DA, Kuska MI, Kanavalli M, Klimecka MM, Merski M, Bandyra KJ, Górna MW
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
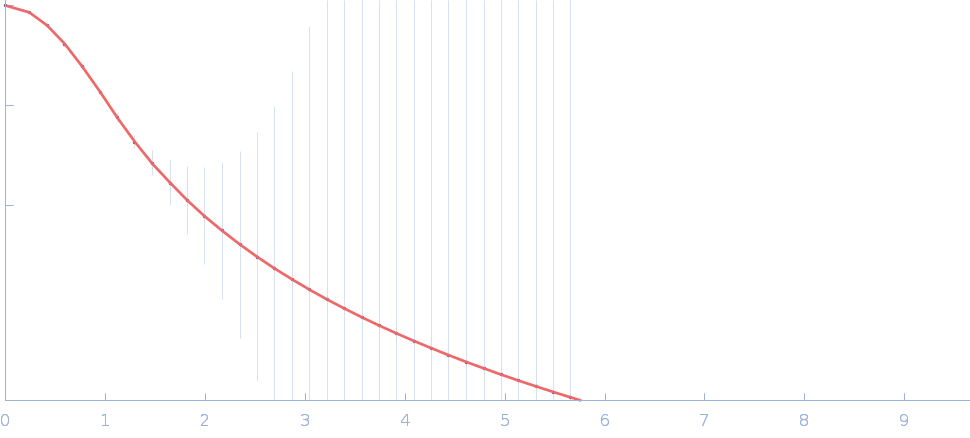
|
| Sample: |
Fas-activated serine/threonine kinase (amino acids 169-549) monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5% (v/v) Glycerol, 200 mM Ammonium Sulphate, 50 mM NaCl, 0.5 mM TCEP, 10 mM Glutamic acid, 10 mM Arginine;, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 31
|
Human FASTK preferentially binds single-stranded and G-rich RNA.
FEBS J (2025)
Dawidziak DM, Dzadz DA, Kuska MI, Kanavalli M, Klimecka MM, Merski M, Bandyra KJ, Górna MW
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Precursor microRNA 20a monomer, 23 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, 5 mM BME, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 8
|
Structural features within precursor microRNA-20a regulate Dicer-TRBP processing
Journal of Molecular Biology :169317 (2025)
Liu Y, Harkner C, Westwood M, Munsayac A, Keane S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
500mM KNO3, 20mM HEPES, 5% glycerol, 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Oct 22
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
7.9 |
nm |
| Dmax |
42.5 |
nm |
| VolumePorod |
485 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing Factor Proline/Glutamine rich (1-598) dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
500mM KNO3, 20mM HEPES, 5% glycerol, 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 13
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
7.6 |
nm |
| Dmax |
34.3 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing Factor Proline/Glutamine rich (1-598) dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
150 mM KCl, 20 mM HEPES, 5% glycerol, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 13
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
6.1 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
315 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
150 mM KCl, 0.075% glycerol, 20 mM HEPES, 0.5 mM DTT,, pH: 7.4 |
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2023 Jan 20
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
6.1 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
150 mM KCl, 1.5% glycerol, 20 mM HEPES, 1 mM DTT (in 100% H2O), pH: 7.4 |
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2023 Jan 23
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
8.7 |
nm |
| Dmax |
30.7 |
nm |
| VolumePorod |
241000 |
nm3 |
|
|