|
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase DTX3L pentamer, 114 kDa Homo sapiens protein
|
| Buffer: |
30 mM HEPES, 350 mM NaCl, 10% glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 6
|
Reconstitution of the DTX3L-PARP9 complex reveals determinants for high affinity heterodimerization and multimeric assembly.
Biochem J (2022)
Ashok Y, Vela-Rodríguez C, Yang CS, Alanen HI, Liu F, Paschal BM, Lehtiö L
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
200 |
nm3 |
|
|
|
|
|
|
|
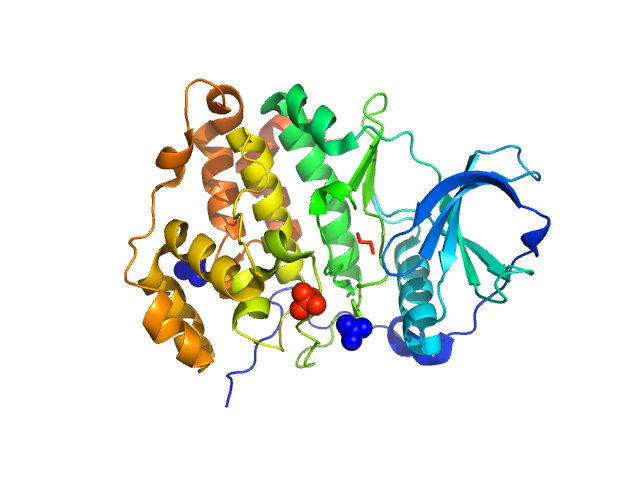
| Sample: |
Anti-CD20 IgG antibody (Rituximab heavy chain chimeric) dimer, 98 kDa Homo sapiens protein
Anti-CD20 IgG antibody (Rituximab light chain chimeric) dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, pH: 7.7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 20
|
Structural Characterization of the Full-Length Anti-CD20 Antibody Rituximab.
Front Mol Biosci 9:823174 (2022)
Belviso BD, Mangiatordi GF, Alberga D, Mangini V, Carrozzini B, Caliandro R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
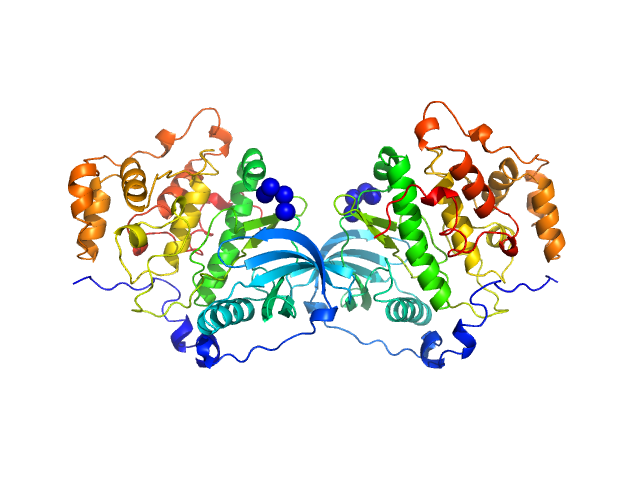
| Sample: |
Casein kinase II subunit alpha monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 500 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Oct 10
|
Mechanism of CK2 Inhibition by a Ruthenium-Based Polyoxometalate.
Front Mol Biosci 9:906390 (2022)
Fabbian S, Giachin G, Bellanda M, Borgo C, Ruzzene M, Spuri G, Campofelice A, Veneziano L, Bonchio M, Carraro M, Battistutta R
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
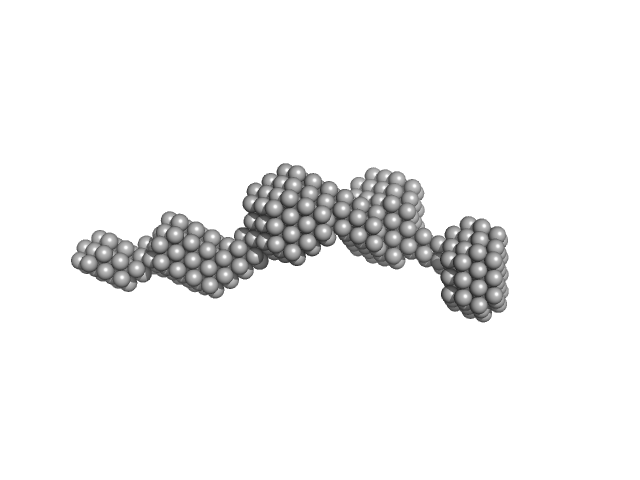
| Sample: |
Casein kinase II subunit alpha dimer, 78 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 500 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Oct 10
|
Mechanism of CK2 Inhibition by a Ruthenium-Based Polyoxometalate.
Front Mol Biosci 9:906390 (2022)
Fabbian S, Giachin G, Bellanda M, Borgo C, Ruzzene M, Spuri G, Campofelice A, Veneziano L, Bonchio M, Carraro M, Battistutta R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Centrosomal-P4.1-associated-protein monomer, 12 kDa Homo sapiens protein
Centriolar coiled-coil protein of 110 kDa dimer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 23
|
Centriolar cap proteins CP110 and CPAP control slow elongation of microtubule plus ends.
J Cell Biol 224(3) (2025)
Iyer SS, Chen F, Ogunmolu FE, Moradi S, Volkov VA, van Grinsven EJ, van Hoorn C, Wu J, Andrea N, Hua S, Jiang K, Vakonakis I, Potočnjak M, Herzog F, Gigant B, Gudimchuk N, Stecker KE, Dogterom M, Steinmetz MO, Akhmanova A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
35 |
nm3 |
|
|