|
|
|
|
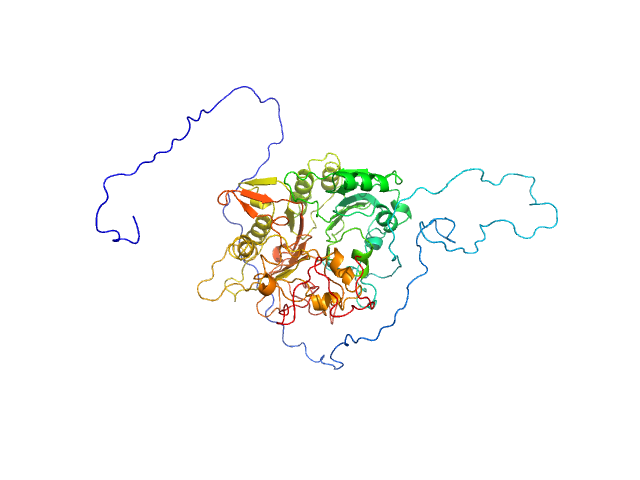
|
| Sample: |
Tyrosyl-DNA phosphodiesterase 1 (dephosphorylated) monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 26
|
Direct interaction of DNA repair protein tyrosyl DNA phosphodiesterase 1 and the DNA ligase III catalytic domain is regulated by phosphorylation of its flexible N-terminus.
J Biol Chem :100921 (2021)
Rashid I, Hammel M, Sverzhinsky A, Tsai MS, Pascal JM, Tainer JA, Tomkinson AE
|
| RgGuinier |
3.2 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
|
|
|
|
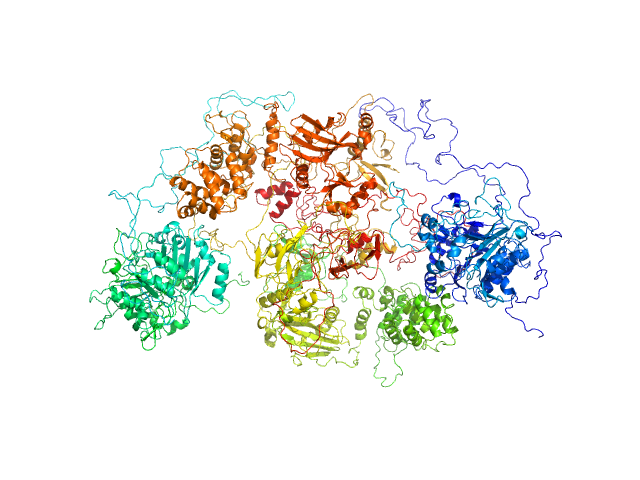
|
| Sample: |
DNA ligase 3 (DNA ligase III alpha), Homo sapiens protein
Tyrosyl-DNA phosphodiesterase 1 monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 40 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
Direct interaction of DNA repair protein tyrosyl DNA phosphodiesterase 1 and the DNA ligase III catalytic domain is regulated by phosphorylation of its flexible N-terminus.
J Biol Chem :100921 (2021)
Rashid I, Hammel M, Sverzhinsky A, Tsai MS, Pascal JM, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.5 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
790 |
nm3 |
|
|
|
|
|
|
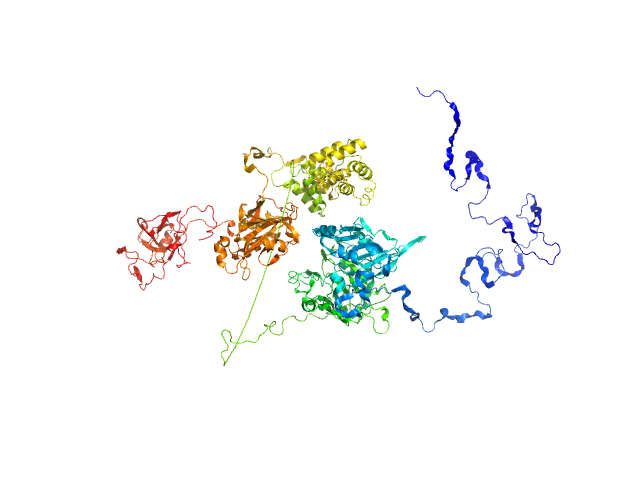
|
| Sample: |
Tyrosyl-DNA phosphodiesterase 1 monomer, 71 kDa Homo sapiens protein
Isoform 3 of DNA ligase 3 (DNA ligase III alpha) monomer, 68 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 40 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jul 5
|
Direct interaction of DNA repair protein tyrosyl DNA phosphodiesterase 1 and the DNA ligase III catalytic domain is regulated by phosphorylation of its flexible N-terminus.
J Biol Chem :100921 (2021)
Rashid I, Hammel M, Sverzhinsky A, Tsai MS, Pascal JM, Tainer JA, Tomkinson AE
|
| RgGuinier |
4.5 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
|
|
|
|
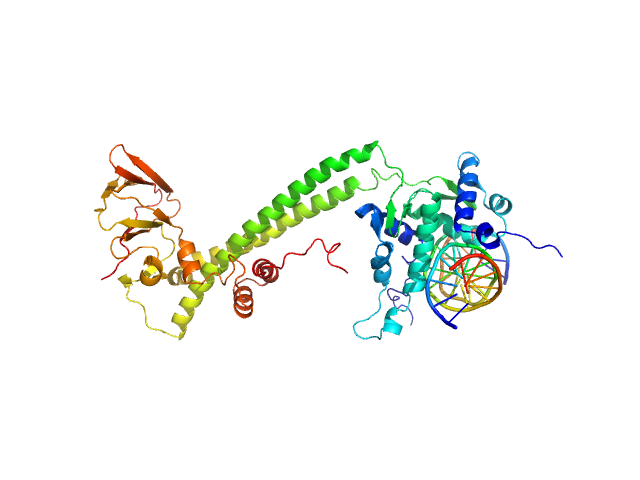
|
| Sample: |
Transcription factor E2F1 monomer, 20 kDa Homo sapiens protein
15-mer DNA oligo of the human E2F1 promoter monomer, 9 kDa DNA
E2F dimerization partner 1 monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-Cl pH 8.0, 150 mM NaCl and 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 19
|
High Conformational Flexibility of the E2F1/DP1/DNA Complex.
J Mol Biol 433(18):167119 (2021)
Saad D, Paissoni C, Chaves-Sanjuan A, Nardini M, Mantovani R, Gnesutta N, Camilloni C
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
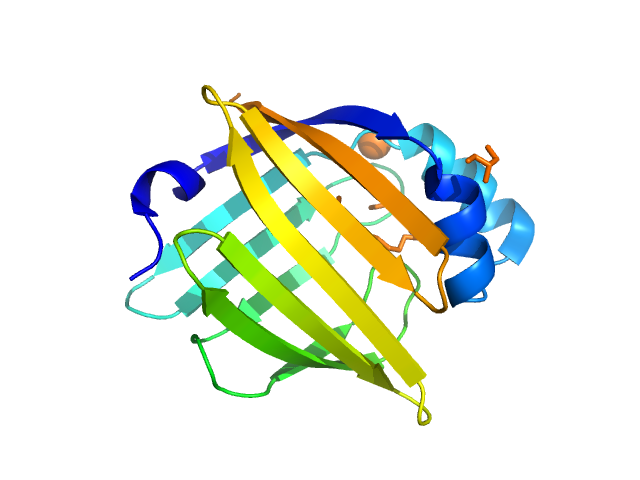
|
| Sample: |
Myelin P2 protein monomer, 15 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jul 22
|
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
FEBS J (2021)
Uusitalo M, Klenow MB, Laulumaa S, Blakeley MP, Simonsen AC, Ruskamo S, Kursula P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.2 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
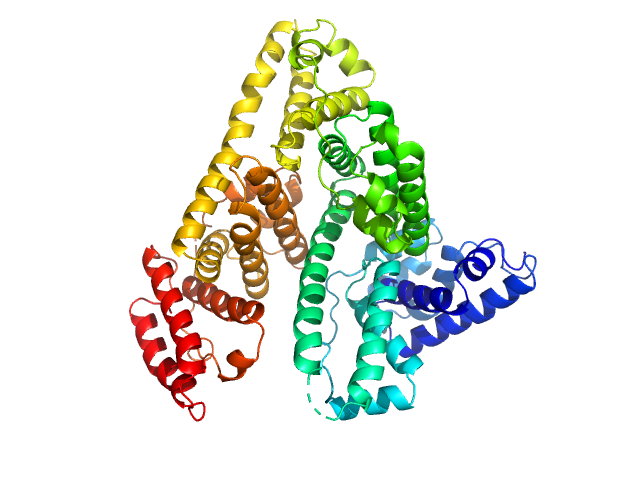
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.4 |
nm |
|
|
|
|
|
|
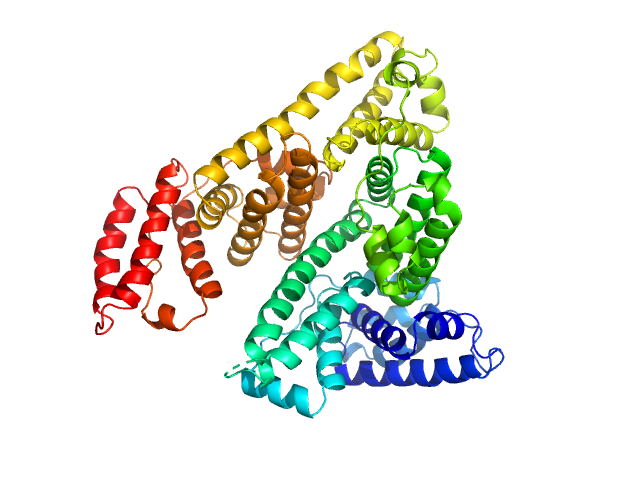
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
|
|