|
|
|
|
|
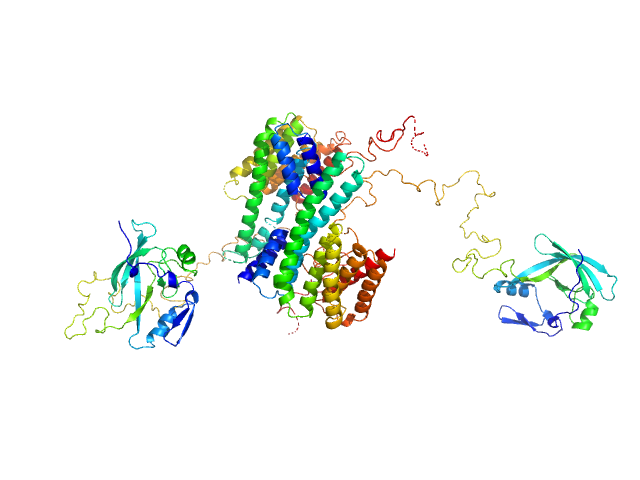
| Sample: |
Ataxin-1 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 13
|
A structural study of the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1.
J Mol Biol :167174 (2021)
Leysen S, Jane Burnley R, Rodriguez E, Milroy LG, Soini L, Adamski CJ, Nitschke L, Davis R, Obsil T, Brunsveld L, Crabbe T, Yahya Zoghbi H, Ottmann C, Martin Davis J
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
14-3-3 protein zeta/delta dimer, 53 kDa Homo sapiens protein
Ataxin-1 AXH-C dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 13
|
A structural study of the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1.
J Mol Biol :167174 (2021)
Leysen S, Jane Burnley R, Rodriguez E, Milroy LG, Soini L, Adamski CJ, Nitschke L, Davis R, Obsil T, Brunsveld L, Crabbe T, Yahya Zoghbi H, Ottmann C, Martin Davis J
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
14-3-3 protein zeta/delta dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 13
|
A structural study of the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1.
J Mol Biol :167174 (2021)
Leysen S, Jane Burnley R, Rodriguez E, Milroy LG, Soini L, Adamski CJ, Nitschke L, Davis R, Obsil T, Brunsveld L, Crabbe T, Yahya Zoghbi H, Ottmann C, Martin Davis J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
7.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
Syndecan-1 monomer, 27 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 26
|
Extended disorder at the cell surface: the conformational landscape of the ectodomains of syndecans
Matrix Biology Plus :100081 (2021)
Gondelaud F, Bouakil M, Le Fèvre A, Erica Miele A, Chirot F, Duclos B, Liwo A, Ricard-Blum S
|
|
|
|
|
|
|
|
| Sample: |
Syndecan-2 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 31
|
Extended disorder at the cell surface: the conformational landscape of the ectodomains of syndecans
Matrix Biology Plus :100081 (2021)
Gondelaud F, Bouakil M, Le Fèvre A, Erica Miele A, Chirot F, Duclos B, Liwo A, Ricard-Blum S
|
|
|
|
|
|
|
|
| Sample: |
Syndecan-3 monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 May 29
|
Extended disorder at the cell surface: the conformational landscape of the ectodomains of syndecans
Matrix Biology Plus :100081 (2021)
Gondelaud F, Bouakil M, Le Fèvre A, Erica Miele A, Chirot F, Duclos B, Liwo A, Ricard-Blum S
|
|
|
|
|
|
|
|
| Sample: |
Syndecan-4 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Jul 4
|
Extended disorder at the cell surface: the conformational landscape of the ectodomains of syndecans
Matrix Biology Plus :100081 (2021)
Gondelaud F, Bouakil M, Le Fèvre A, Erica Miele A, Chirot F, Duclos B, Liwo A, Ricard-Blum S
|
|
|
|
|
|
|
|
| Sample: |
Syndecan-4 dimer, 36 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Jul 4
|
Extended disorder at the cell surface: the conformational landscape of the ectodomains of syndecans
Matrix Biology Plus :100081 (2021)
Gondelaud F, Bouakil M, Le Fèvre A, Erica Miele A, Chirot F, Duclos B, Liwo A, Ricard-Blum S
|
|
|
|
|
|
|
|
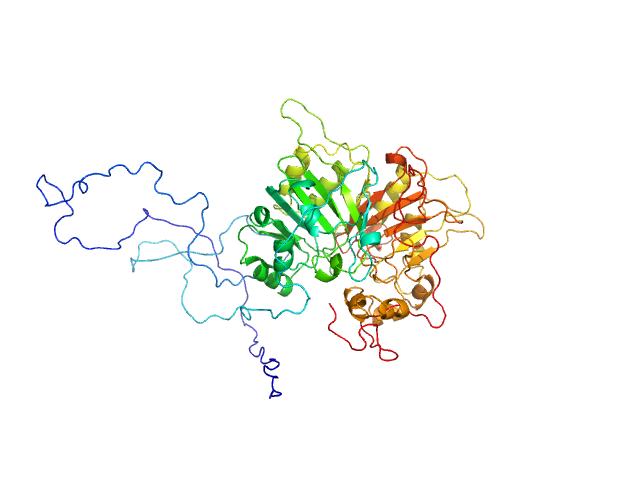
| Sample: |
Tyrosyl-DNA phosphodiesterase 1 monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 26
|
Direct interaction of DNA repair protein tyrosyl DNA phosphodiesterase 1 and the DNA ligase III catalytic domain is regulated by phosphorylation of its flexible N-terminus.
J Biol Chem :100921 (2021)
Rashid I, Hammel M, Sverzhinsky A, Tsai MS, Pascal JM, Tainer JA, Tomkinson AE
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
|
|
|
|
|
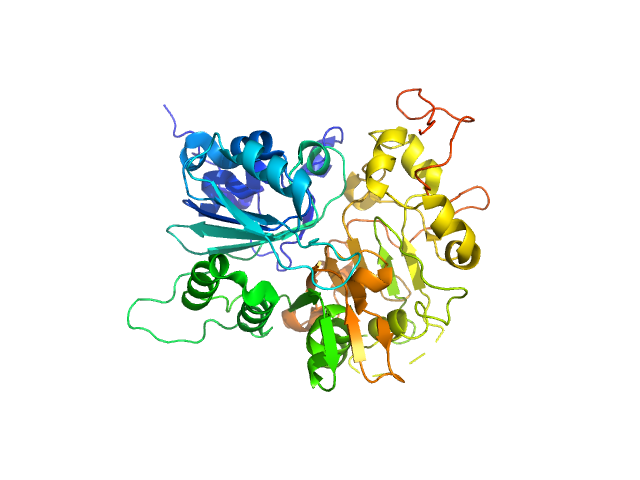
| Sample: |
Tyrosyl-DNA phosphodiesterase 1 (149-608) monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 18
|
Direct interaction of DNA repair protein tyrosyl DNA phosphodiesterase 1 and the DNA ligase III catalytic domain is regulated by phosphorylation of its flexible N-terminus.
J Biol Chem :100921 (2021)
Rashid I, Hammel M, Sverzhinsky A, Tsai MS, Pascal JM, Tainer JA, Tomkinson AE
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|