|
|
|
|
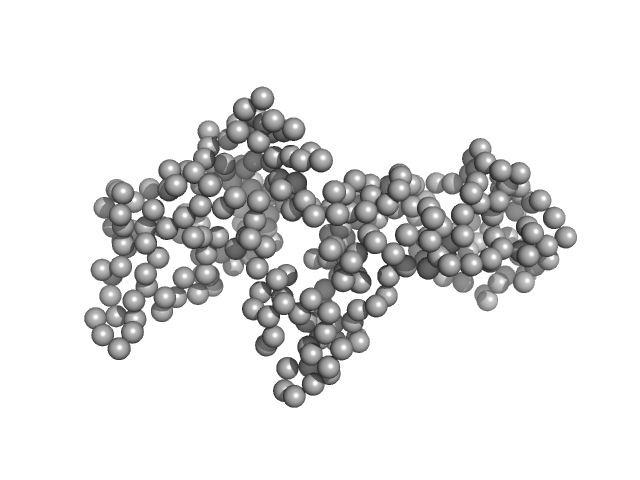
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆295-358 monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
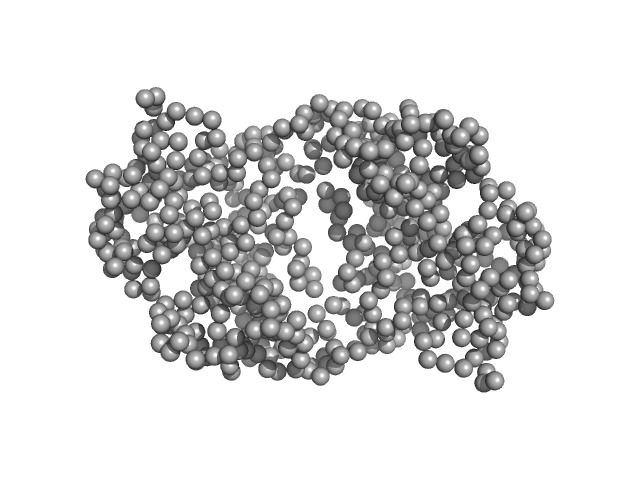
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆303-358 dimer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 28
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆303-358, mutant Y29E/C88A monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 28
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358 dimer, 74 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
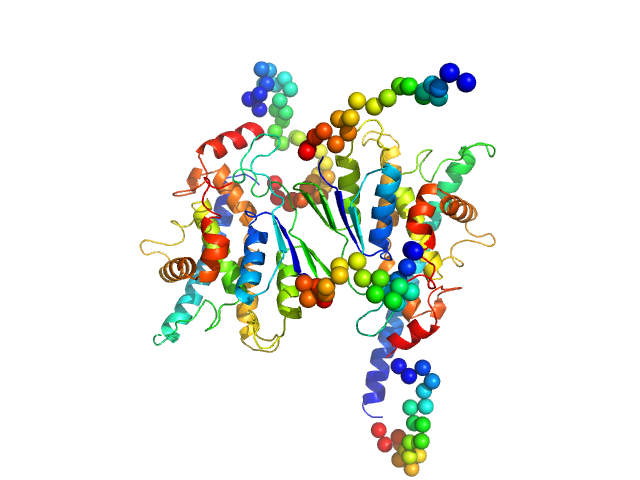
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358 dimer, 74 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|
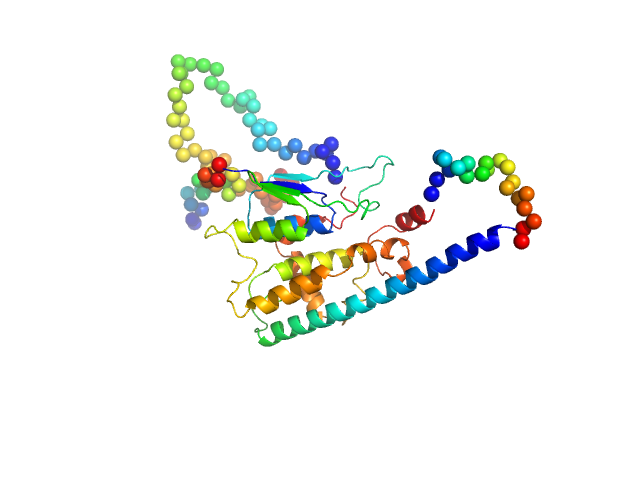
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1-like 1 (∆125-143 isoform) monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM TRIS pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 10
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKA4_fit1_model1.png)
|
| Sample: |
Properdin (dimer) dimer, 110 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
8.1 |
nm |
| Dmax |
24.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKB4_fit1_model1.png)
|
| Sample: |
Properdin (trimer) trimer, 165 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
10.2 |
nm |
| Dmax |
27.0 |
nm |
|
|