|
|
|
|
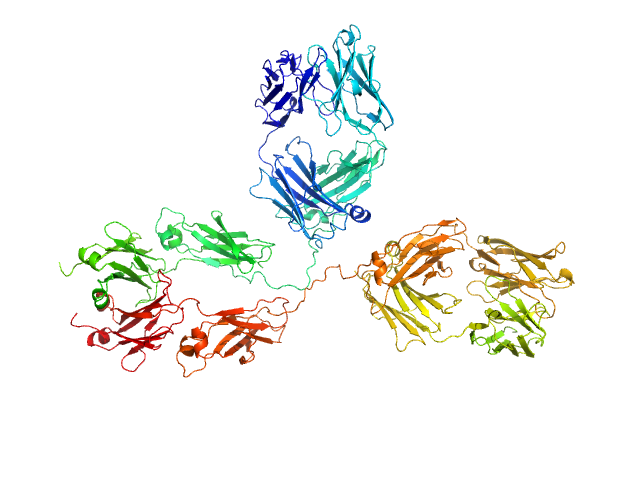
|
| Sample: |
Immunoglobulin G subclass 1, 148 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 20
|
Solution structure of deglycosylated human IgG1 shows the role of CH2 glycans in its conformation
Biophysical Journal (2021)
Spiteri V, Doutch J, Rambo R, Gor J, Dalby P, Perkins S
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
271 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDK68_fit1_model1.png)
|
| Sample: |
Nuclear receptor coactivator 2 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 50 mM BisTris pH 6.8, and 0.5 mM EDTA, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 14
|
Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J Mol Biol 433(9):166899 (2021)
Senicourt L, le Maire A, Allemand F, Carvalho JE, Guee L, Germain P, Schubert M, Bernadó P, Bourguet W, Sibille N
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
TC1 monomer, 3 kDa synthetic construct DNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Jun 26
|
Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1.
Nucleic Acids Res (2021)
Loughlin FE, West DL, Gunzburg MJ, Waris S, Crawford SA, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
UC1 monomer, 3 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Jun 26
|
Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1.
Nucleic Acids Res (2021)
Loughlin FE, West DL, Gunzburg MJ, Waris S, Crawford SA, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Modified Nucleolysin TIA-1 isofrom p40 monomer, 42 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 60 mM KCl, 0.5 M arginine-HCl, 1 mM MgCl2, 2 mM DTT, 0.5 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2020 Jul 7
|
Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1.
Nucleic Acids Res (2021)
Loughlin FE, West DL, Gunzburg MJ, Waris S, Crawford SA, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
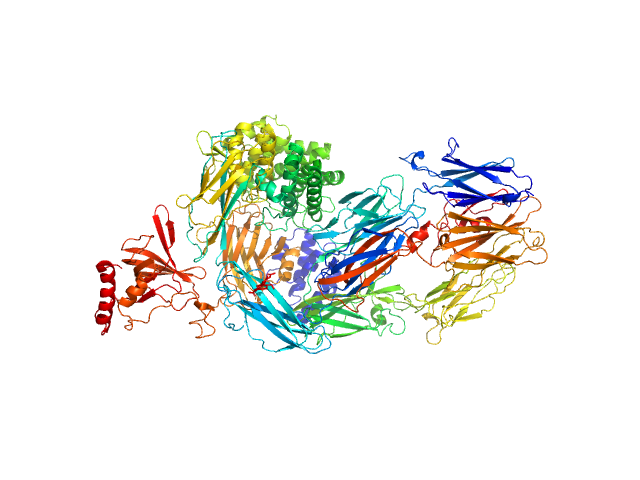
|
| Sample: |
Complement C5 monomer, 186 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris pH, 75mM NaCl, and 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 12
|
The allosteric modulation of Complement C5 by knob domain peptides.
Elife 10 (2021)
Macpherson A, Laabei M, Ahdash Z, Graewert MA, Birtley JR, Schulze ME, Crennell S, Robinson SA, Holmes B, Oleinikovas V, Nilsson PH, Snowden J, Ellis V, Mollnes TE, Deane CM, Svergun D, Lawson AD, van den Elsen JM
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
392 |
nm3 |
|
|
|
|
|
|
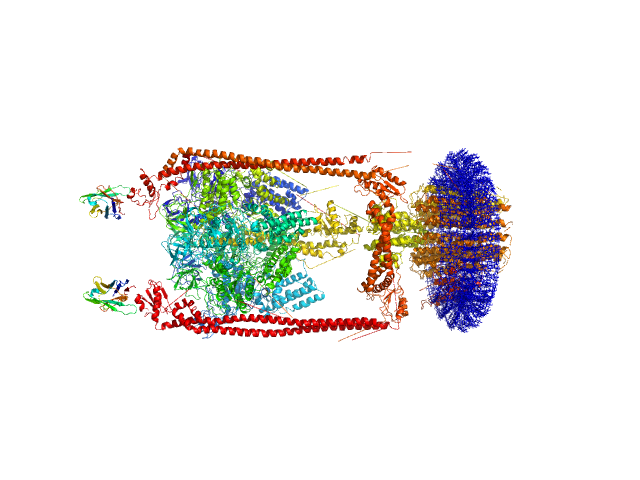
|
| Sample: |
N-Dodecyl-β-D-Maltopyranoside, 77 kDa synthetic construct
A-type ATP synthase monomer, 655 kDa Thermus thermophilus protein
Monoclonal Antibody Fragment dimer, 33 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris/HCl, 100 mM sucrose, 100 mM NaCl, 2 mM MgCl2, 10% glycerol, 0.05% b-DDM, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 Mar 3
|
MPBuilder: A PyMOL Plugin for Building and Refinement of Solubilized Membrane Proteins Against Small Angle X-ray Scattering Data
Journal of Molecular Biology :166888 (2021)
Molodenskiy D, Svergun D, Mertens H
|
| RgGuinier |
8.0 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
1549 |
nm3 |
|
|
|
|
|
|
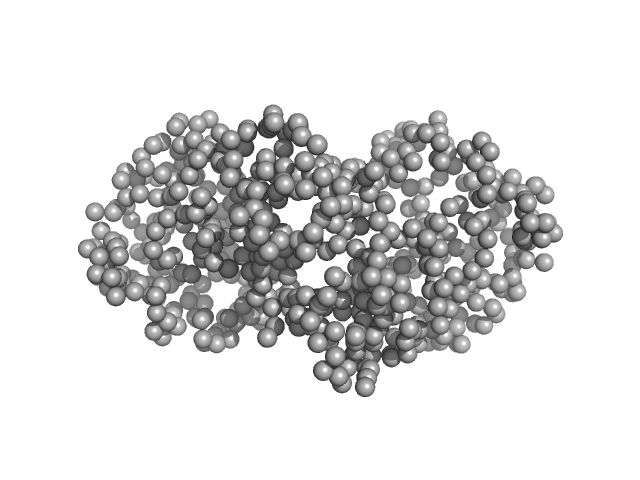
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆295-358 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 21
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
101.6 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆295-358 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆295-358 monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 21
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
92.3 |
nm |
| VolumePorod |
71 |
nm3 |
|
|