|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
25-mer dsRNA monomer, 16 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
431 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
36-mer dsRNA monomer, 23 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
587 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
36-mer dsRNA monomer, 23 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.1 |
nm |
| VolumePorod |
626 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.1 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
797 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.7 |
nm |
| Dmax |
26.5 |
nm |
| VolumePorod |
1075 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.8 |
nm |
| Dmax |
26.1 |
nm |
| VolumePorod |
1050 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fc fragment of IgG binding protein dimer, 552 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 10 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 20
|
The structure of FCGBP is formed as a disulfide-mediated homodimer between its C-terminal domains.
FEBS J (2025)
Ehrencrona E, Gallego P, Trillo-Muyo S, Garcia-Bonete MJ, Recktenwald CV, Hansson GC, Johansson MEV
|
| RgGuinier |
10.0 |
nm |
| Dmax |
42.0 |
nm |
| VolumePorod |
1462 |
nm3 |
|
|
|
|
|
|
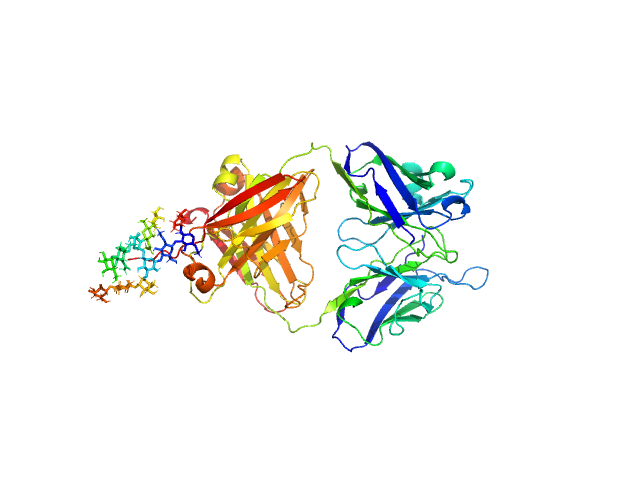
|
| Sample: |
IgM Mannitou Fab Heavy Chain monomer, 26 kDa Mus musculus protein
IgM Mannitou Fab Light Chain monomer, 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Jun 5
|
Small-angle X-ray scattering of engineered antigen-binding fragments: the case of glycosylated Fab from the Mannitou IgM antibody.
Acta Crystallogr F Struct Biol Commun (2025)
Semwal S, Karamolegkou M, Flament S, Raouraoua N, Verstraete K, Thureau A, Wien F, Bray F, Savvides SN, Bouckaert J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Type 2 DNA topoisomerase 6 subunit B-like monomer, 61 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 500 mM KCl, 5% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 12
|
The TOPOVIBL meiotic DSB formation protein: new insights from its biochemical and structural characterization
Nucleic Acids Research (2024)
Diagouraga B, Tambones I, Carivenc C, Bechara C, Nadal M, de Massy B, le Maire A, Robert T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alarmin release inhibitor (Δ1-62) monomer, 26 kDa Heligmosomoides polygyrus protein
Interleukin-33 (L179V) monomer, 18 kDa Mus musculus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|