|
|
|
|
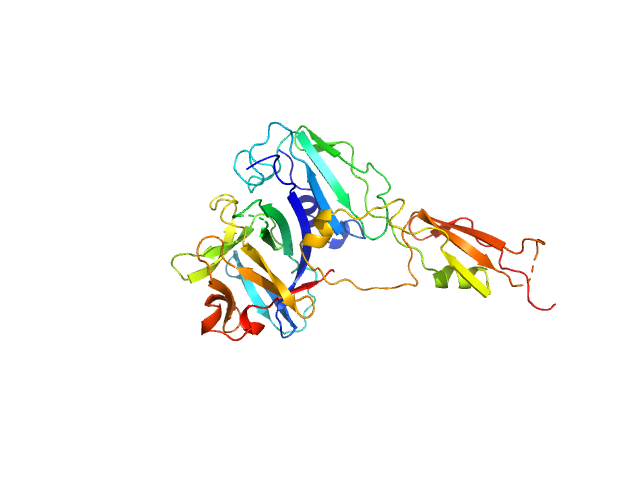
|
| Sample: |
Interleukin-33 (L179V) monomer, 18 kDa Mus musculus protein
Alarmin release inhibitor (Δ1-125; N175Q, N190Q) monomer, 19 kDa Heligmosomoides polygyrus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.9 |
nm |
| Dmax |
37.0 |
nm |
| VolumePorod |
1059 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
1133 |
nm3 |
|
|
|
|
|
|
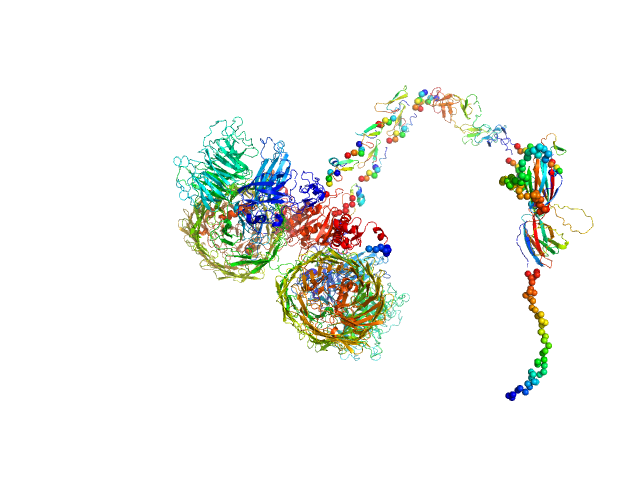
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
1129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM EDTA, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.8 |
nm |
| Dmax |
35.0 |
nm |
| VolumePorod |
1035 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
9.4 |
nm |
| Dmax |
39.0 |
nm |
| VolumePorod |
1101 |
nm3 |
|
|
|
|
|
|
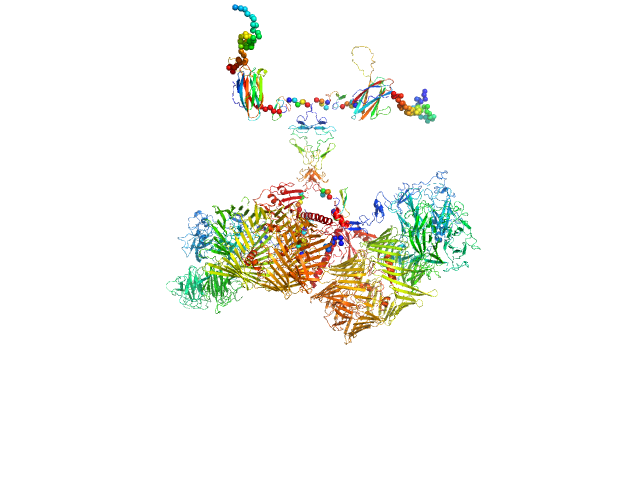
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1057 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1077 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM EDTA, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.6 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
990 |
nm3 |
|
|