|
|
|
|
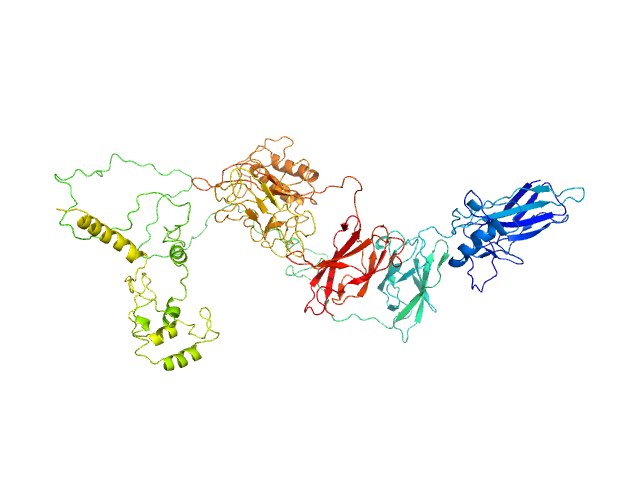
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
|
|
|
|
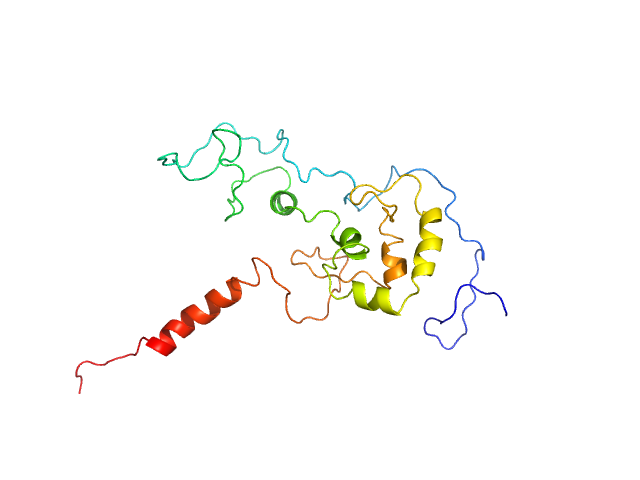
|
| Sample: |
Transcription factor p65 340-549 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
IFN kB DNA monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
Urokinase kB monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
IFN kB DNA monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 4
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
314 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Urokinase kB monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 4
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Roquin-1 monomer, 18 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Roquin-1 extROQ monomer, 35 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
2.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Roquin-1 Nterm monomer, 51 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Roquin-1 Nterm Amut (K220A, K239A, R260A) monomer, 51 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, 2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|