|
|
|
|
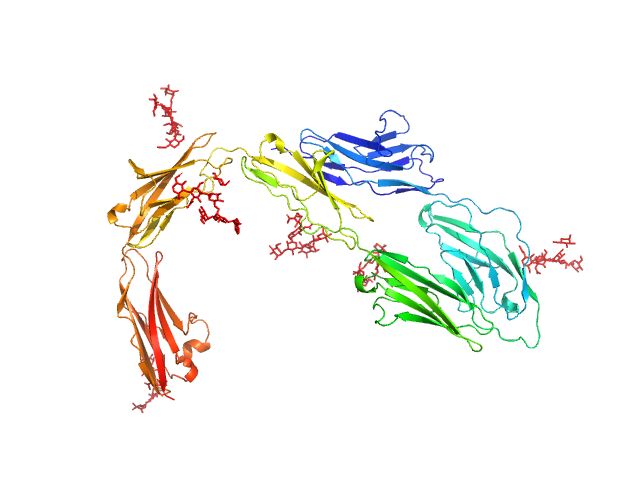
|
| Sample: |
Contactin-1 I433V dimer, 134 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
4.9 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
8.1 |
nm |
| Dmax |
42.0 |
nm |
| VolumePorod |
520 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
8.0 |
nm |
| Dmax |
43.0 |
nm |
| VolumePorod |
530 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
8.0 |
nm |
| Dmax |
43.0 |
nm |
| VolumePorod |
530 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
8.0 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
7.0 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
285 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
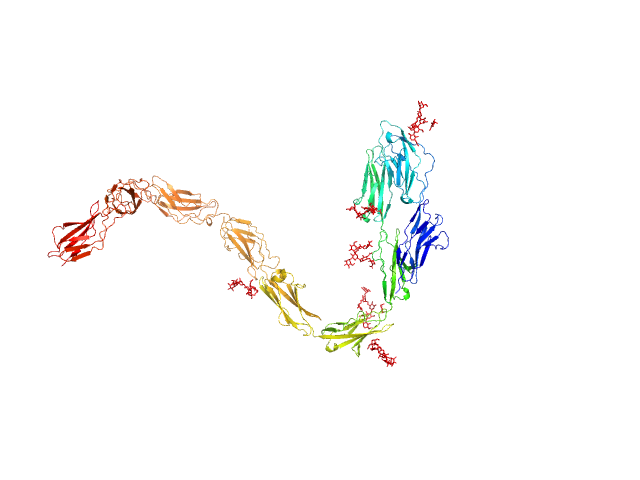
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
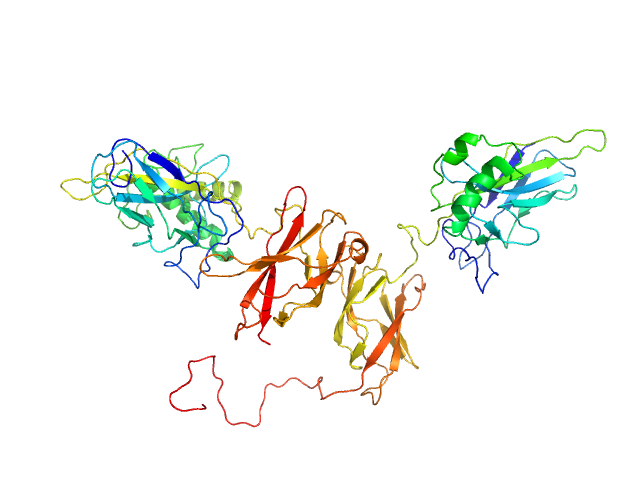
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|