|
|
|
|
|
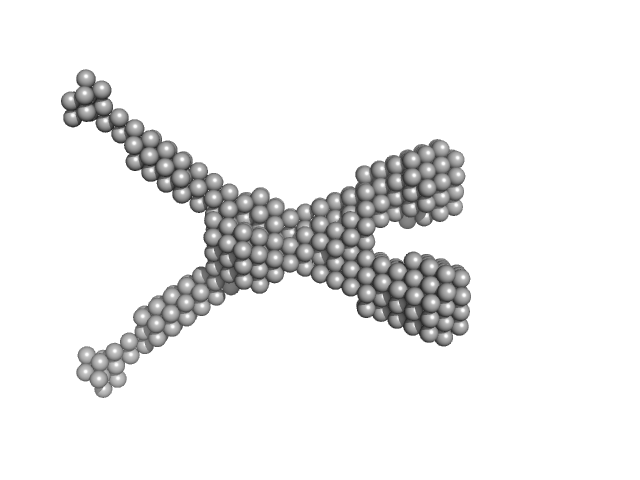
| Sample: |
Noelin tetramer, 72 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol 20(1):50 (2019)
Pronker MF, van den Hoek H, Janssen BJC
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytohesin-3 dimer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|

|
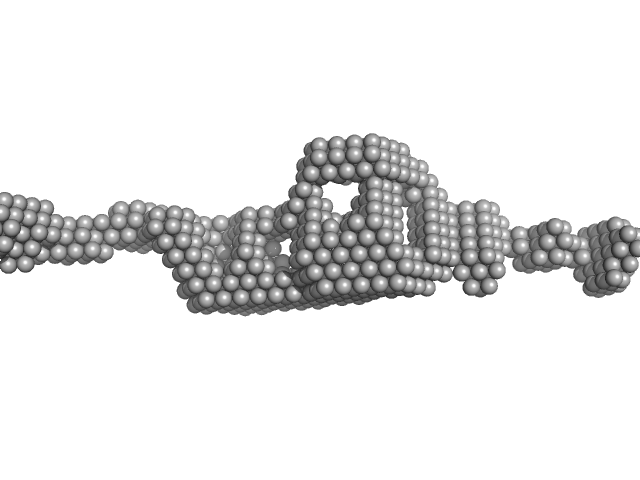
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.3 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
|
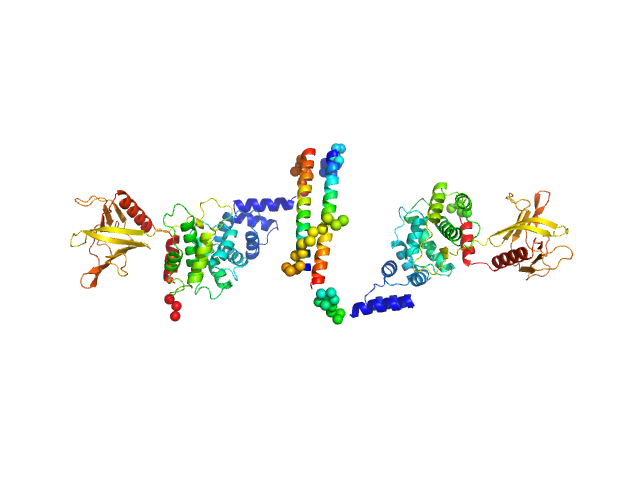
| Sample: |
Cytohesin-2 dimer, 95 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.3 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
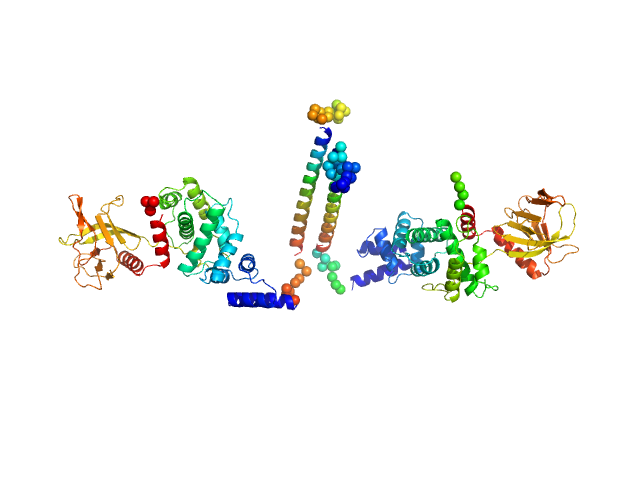
| Sample: |
Cytohesin-3 dimer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|
|
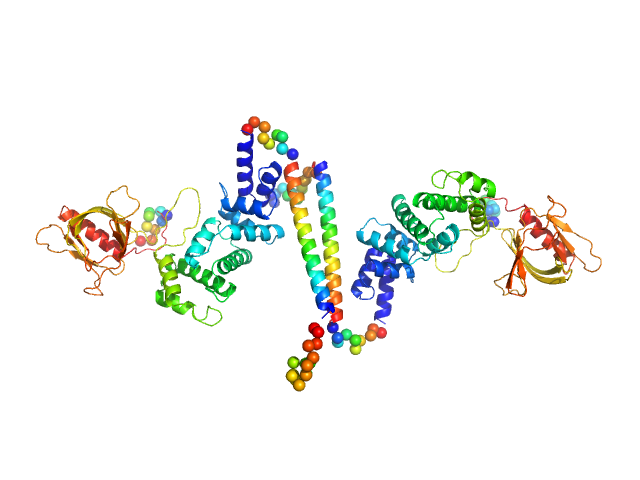
| Sample: |
Cytohesin-3 dimer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|
|
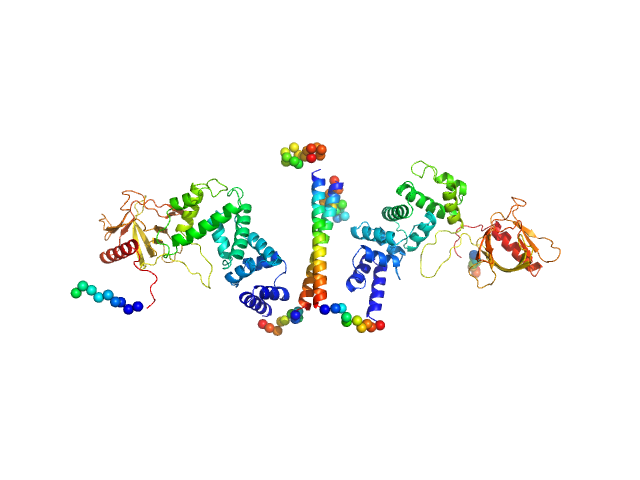
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.1 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.1 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
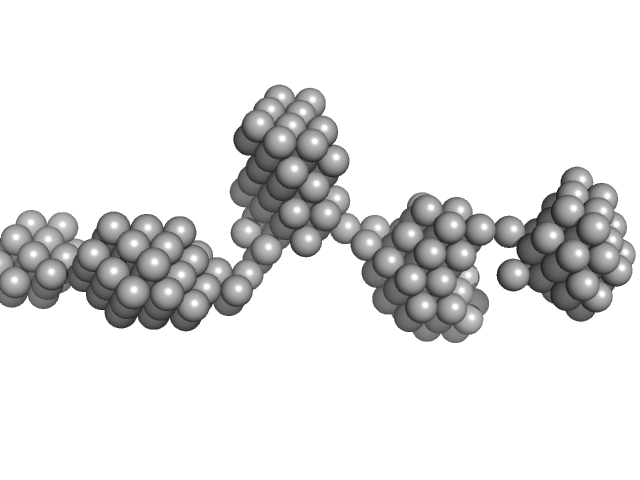
|
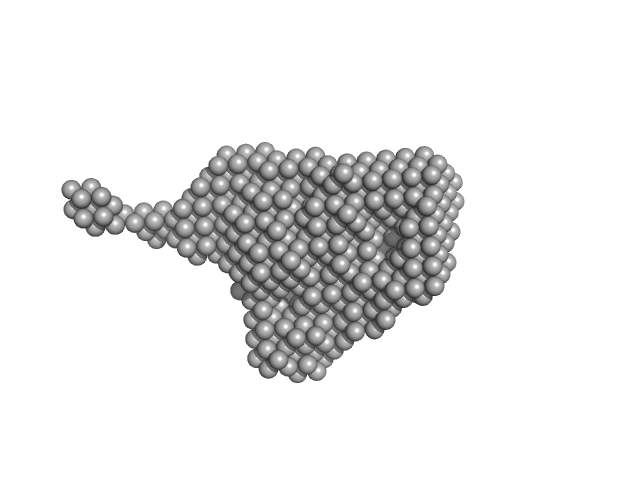
| Sample: |
Aryl hydrocarbon receptor nuclear translocator-like protein 1 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP), mutation L664C monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
1.9 |
nm |
| Dmax |
8.1 |
nm |
|
|