|
|
|
|
|
| Sample: |
Collagen, type VI, alpha 3 monomer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM TRIS, 150mM NaCl 3% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 4
|
Structure of a collagen VI α3 chain VWA domain array: adaptability and functional implications of myopathy causing mutations
Journal of Biological Chemistry :jbc.RA120.014865 (2020)
Solomon-Degefa H, Gebauer J, Jeffries C, Freiburg C, Meckelburg P, Bird L, Baumann U, Svergun D, Owens R, Werner J, Behrmann E, Paulsson M, Wagener R
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
BRME1 dimer, 23 kDa Mus musculus protein
Meiotic localizer of BRCA2 dimer, 26 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Meiotic localizer of BRCA2 dimer, 26 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|

|
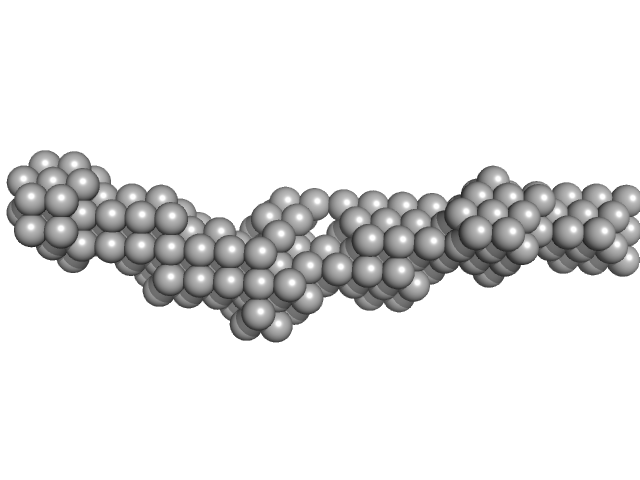
| Sample: |
Meiotic localizer of BRCA2 octamer, 102 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
368 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Meiotic localizer of BRCA2 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|

|
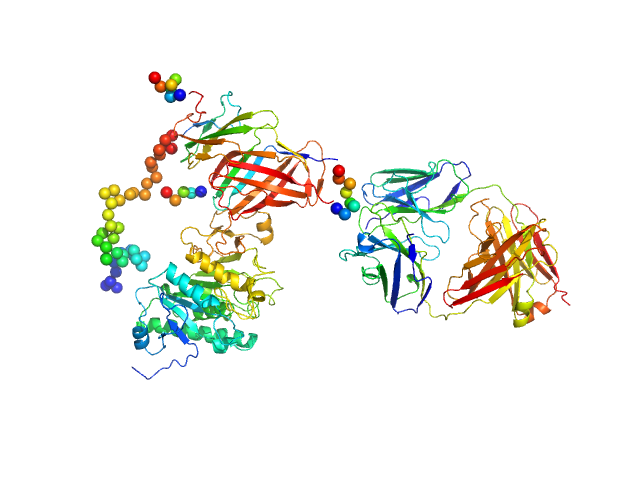
| Sample: |
TRAF-interacting protein with FHA domain-containing protein A dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 100 mM arginine, 5 % glycerol, 10 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Kumamoto University on 2014 Oct 16
|
Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep 10(1):5152 (2020)
Nakamura T, Hashikawa C, Okabe K, Yokote Y, Chirifu M, Toma-Fukai S, Nakamura N, Matsuo M, Kamikariya M, Okamoto Y, Gohda J, Akiyama T, Semba K, Ikemizu S, Otsuka M, Inoue JI, Yamagata Y
|
| RgGuinier |
3.1 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
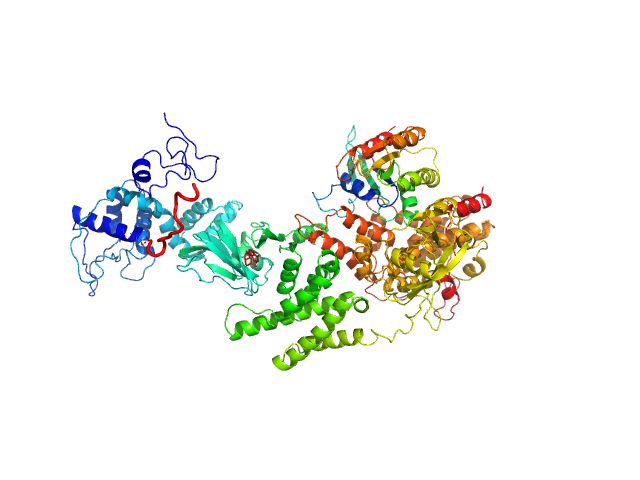
| Sample: |
Lipoprotein lipase monomer, 50 kDa Homo sapiens protein
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 monomer, 15 kDa Homo sapiens protein
Monoclonal Antibody Fragment 5D2 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, 4 mM CaCL2, 10% (v/v) Glycerol, 0.05% 0.8mM CHAPS, ,0.05 % (v/v) NaN3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 14
|
Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism
Proceedings of the National Academy of Sciences :201920202 (2020)
Kristensen K, Leth-Espensen K, Mertens H, Birrane G, Meiyappan M, Olivecrona G, Jørgensen T, Young S, Ploug M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
|
|
|
|
|
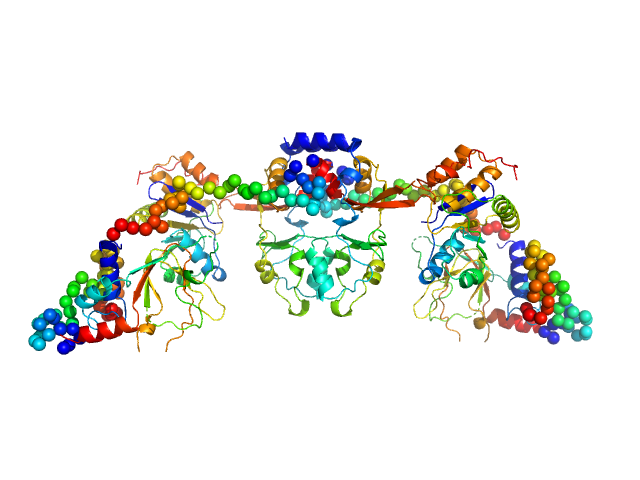
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 100 kDa Homo sapiens protein
RAS related protein 1b monomer, 18 kDa Mus musculus protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, 1mM cAMP, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2013 Apr 1
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|
|
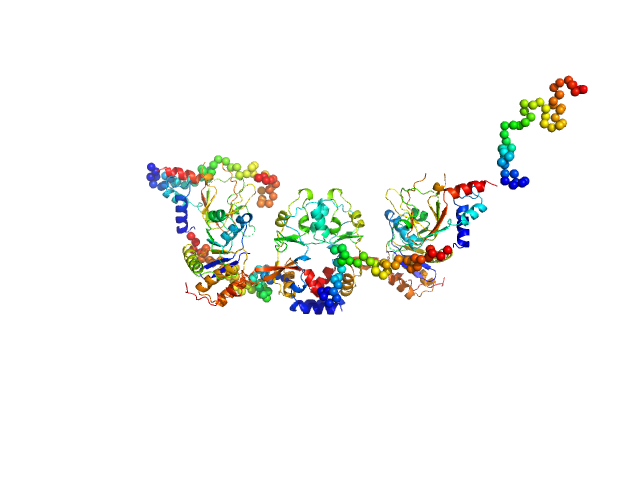
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
Protein tyrosine phosphatase type IVA 1 dimer, 40 kDa Mus musculus protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 17
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks T, Blanco FJ, Díaz Moreno I, Martínez-Chantar ML, Tremblay ML, Müller D, Siliqi D, Martínez-Cruz LA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
216 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
Protein tyrosine phosphatase type IVA 1 dimer, 40 kDa Mus musculus protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Aug 2
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks T, Blanco FJ, Díaz Moreno I, Martínez-Chantar ML, Tremblay ML, Müller D, Siliqi D, Martínez-Cruz LA
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
226 |
nm3 |
|
|