|
|
|
|
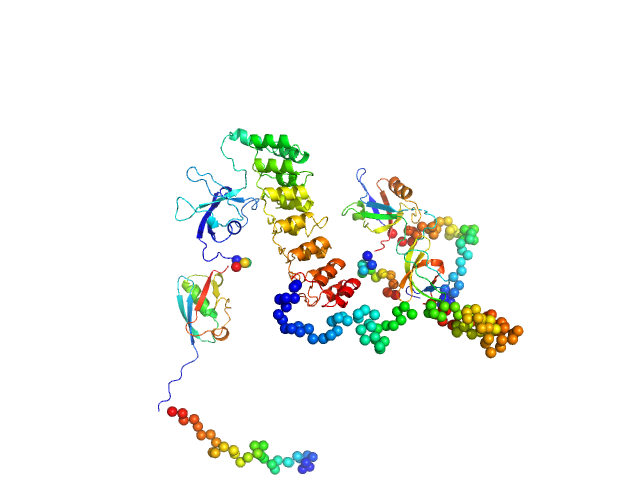
|
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 88 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
|
|
|
|
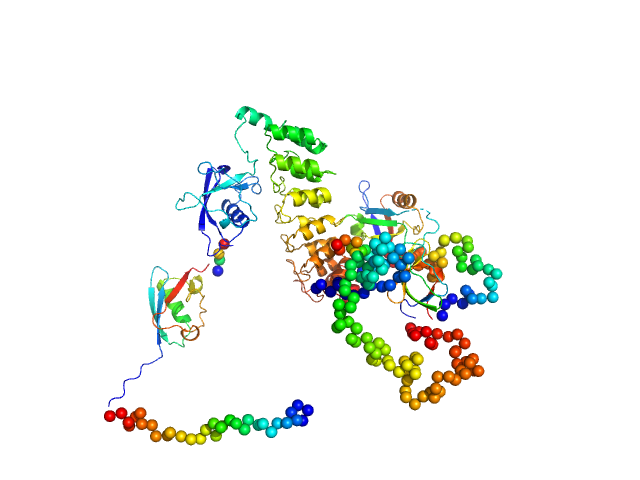
|
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 87 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histone H3 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B monomer, 14 kDa Xenopus laevis protein
Non-linker Ended Trinucleosome DNA monomer, 172 kDa DNA
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 1 mM DTT 50% w/v sucrose, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Mar 11
|
Solution structure(s) of trinucleosomes from contrast variation SAXS
Nucleic Acids Research (2021)
Mauney A, Muthurajan U, Luger K, Pollack L
|
| RgGuinier |
12.9 |
nm |
| Dmax |
41.8 |
nm |
|
|
|
|
|
|
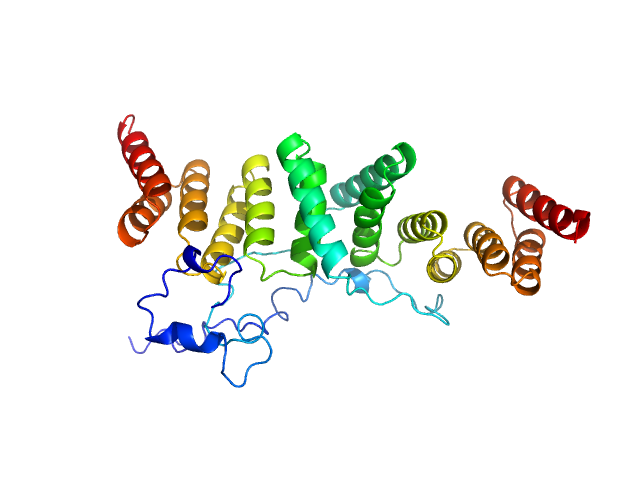
|
| Sample: |
Chaperone protein IpgC dimer, 39 kDa Shigella flexneri protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Dec 10
|
Structural Insights of Shigella Translocator IpaB and Its Chaperone IpgC in Solution
Frontiers in Cellular and Infection Microbiology 11 (2021)
Ferrari M, Charova S, Sansonetti P, Mylonas E, Gazi A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
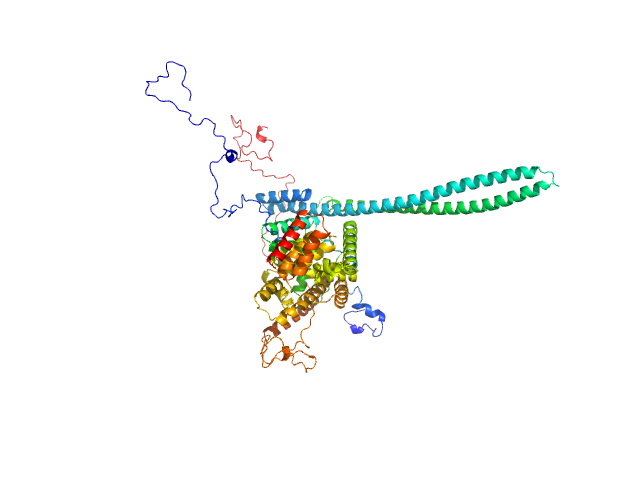
|
| Sample: |
T3SS Chaperone protein IpgC monomer, 20 kDa Shigella flexneri protein
Invasin IpaB monomer, 62 kDa Shigella flexneri protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Dec 10
|
Structural Insights of Shigella Translocator IpaB and Its Chaperone IpgC in Solution
Frontiers in Cellular and Infection Microbiology 11 (2021)
Ferrari M, Charova S, Sansonetti P, Mylonas E, Gazi A
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with RS linker monomer, 23 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with RS linker monomer, 23 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with GGG linker monomer, 23 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with VKA linker monomer, 24 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with AAE linker monomer, 24 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|