|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
1400 mM NaCl, 49.4 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
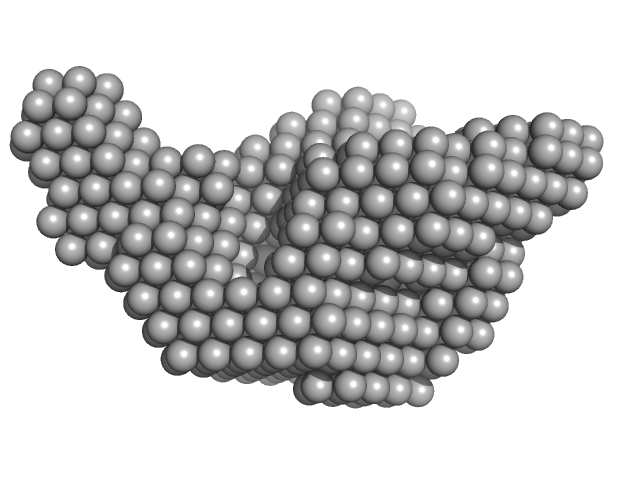
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
9.9 |
nm |
| Dmax |
36.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
2800 mM NaCl, 76.6 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
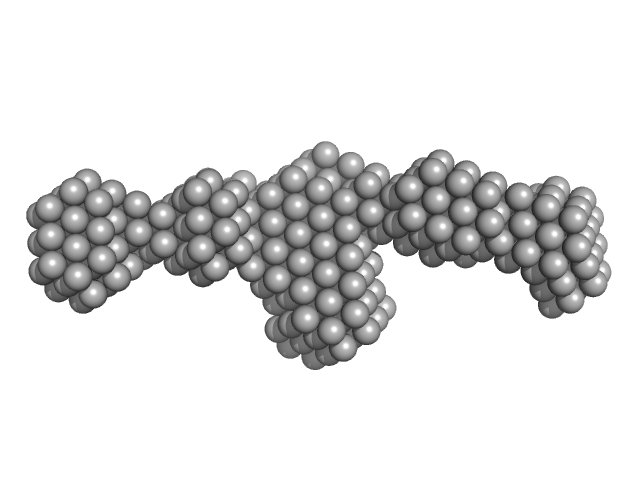
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
9.3 |
nm |
| Dmax |
37.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
4000 mM NaCl, 100 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
8.9 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Signal recognition particle 9 monomer, 12 kDa Plasmodium falciparum protein
Signal recognition particle 14 monomer, 12 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 22
|
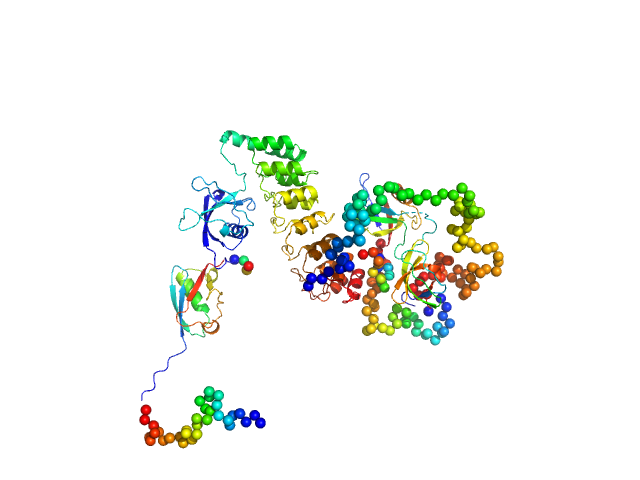
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Full-length SRP Alu RNA monomer, 38 kDa Plasmodium falciparum RNA
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SRP Alu RNA 5' domain monomer, 24 kDa Plasmodium falciparum RNA
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Signal recognition particle 9 monomer, 12 kDa Plasmodium falciparum protein
Signal recognition particle 14 monomer, 12 kDa Plasmodium falciparum protein
Full-length SRP Alu RNA monomer, 38 kDa Plasmodium falciparum RNA
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Signal recognition particle 9 monomer, 12 kDa Plasmodium falciparum protein
Signal recognition particle 14 monomer, 12 kDa Plasmodium falciparum protein
SRP Alu RNA 5' domain monomer, 24 kDa Plasmodium falciparum RNA
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Primer Binding Site-Segment monomer, 33 kDa HIV-1: pNL4-3 RNA
|
| Buffer: |
10 mM Tris, 140 mM KCl, 10 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Jul 29
|
The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.
Nucleic Acids Res (2021)
Song Z, Gremminger T, Singh G, Cheng Y, Li J, Qiu L, Ji J, Lange MJ, Zuo X, Chen SJ, Zou X, Boris-Lawrie K, Heng X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 88 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
170 |
nm3 |
|
|