|
|
|
|

|
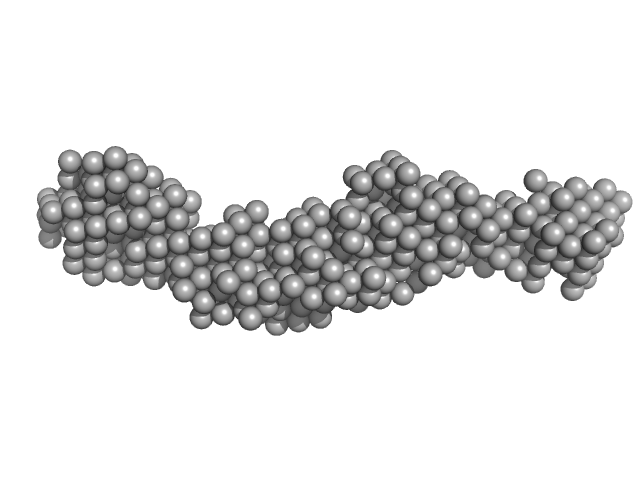
| Sample: |
Collagenase ColH (Full-length) monomer, 112 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 0.4 mM EGTA, 2.4 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 12
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|
|
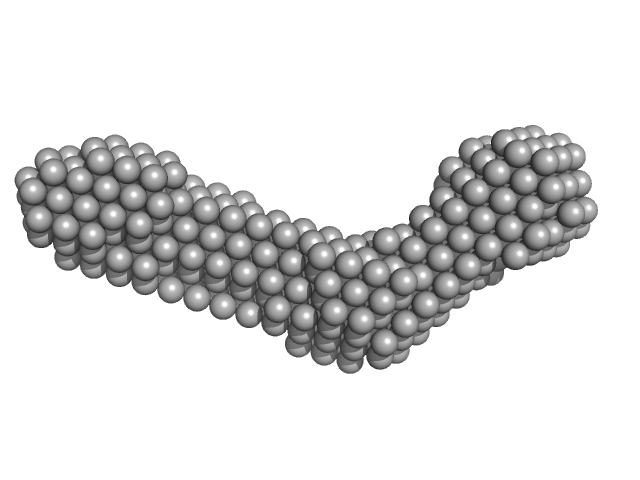
| Sample: |
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr689Ser, Phe712Ser, Tyr780Ser, His782Ser, Tyr796Ser and Tyr801) monomer, 33 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
4.0 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr689Ser, Phe712Ser, Tyr780Ser, His782Ser, Tyr796Ser and Tyr801) monomer, 33 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM Tris, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr689Ser and Phe712Ser) monomer, 33 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 12
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
4.4 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr689Ser and Phe712Ser) monomer, 33 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM Tris, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
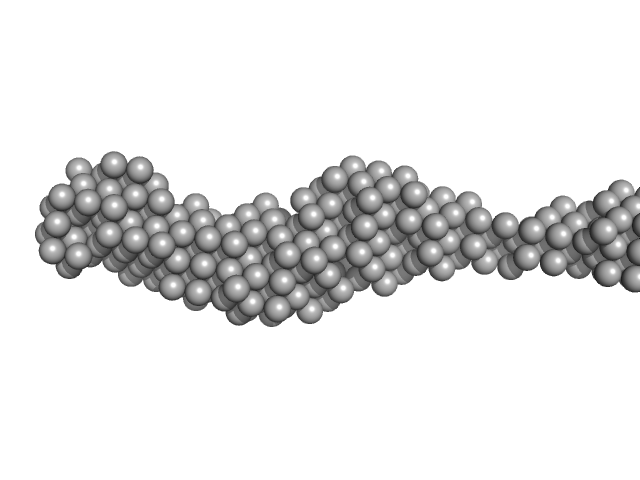
| Sample: |
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD)) monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
4.0 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
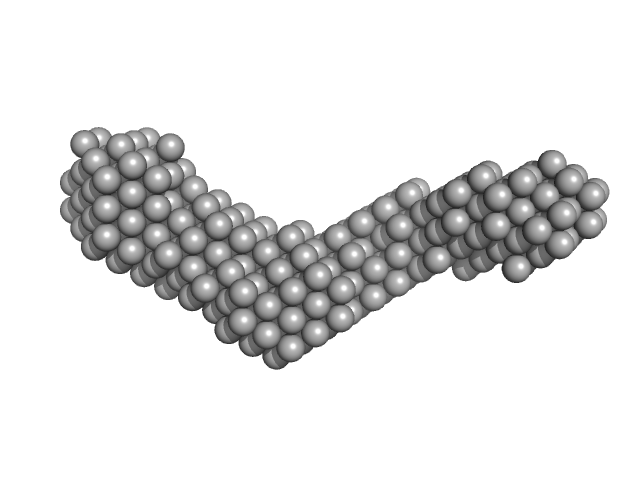
| Sample: |
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD)) monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 0.4 mM EGTA, 2.4 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|

|
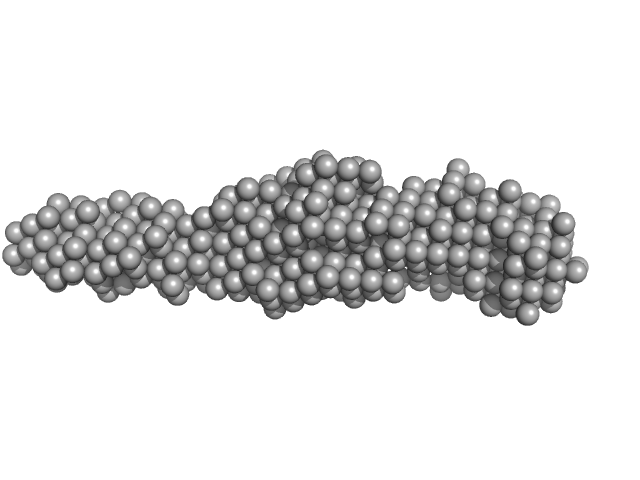
| Sample: |
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr780Ser, His782Ser, Tyr796Ser and Tyr801Ser) monomer, 23 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM Tris, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr780Ser, His782Ser, Tyr796Ser and Tyr801Ser) monomer, 23 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.2 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD)) monomer, 23 kDa Hathewaya histolytica protein
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.3 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
35 |
nm3 |
|
|