|
|
|
|

|
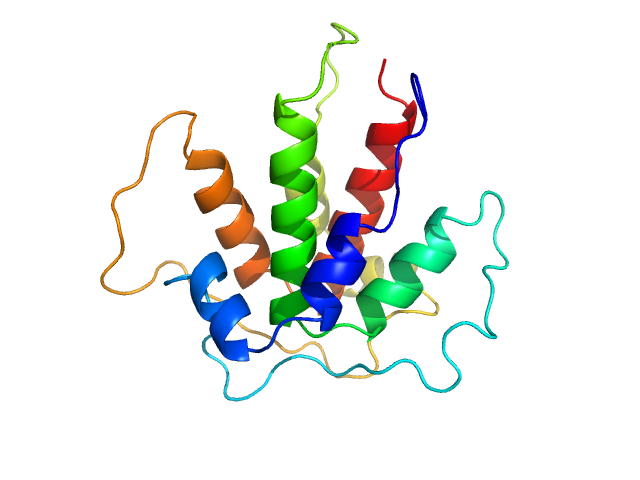
| Sample: |
Beta-ketoacyl synthase Bamb_5920 CDD/Bamb_5919 NDD artificial protein fusion dimer, 13 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
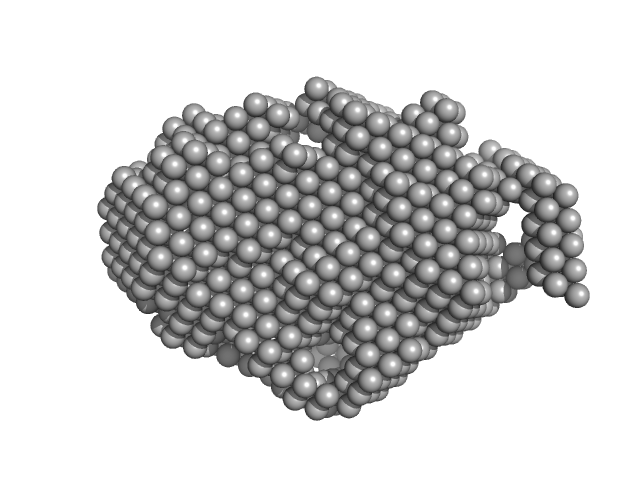
| Sample: |
Beta-ketoacyl synthase Bamb_5925 CDD/Bamb_5924 NDD artificial protein fusion dimer, 18 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, 1 mM trehalose, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, 1 mM UDP-glucose, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 29 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 15
|
Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep 32(3):107930 (2020)
Hollmann NM, Jagtap PKA, Masiewicz P, Guitart T, Simon B, Provaznik J, Stein F, Haberkant P, Sweetapple LJ, Villacorta L, Mooijman D, Benes V, Savitski MM, Gebauer F, Hennig J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
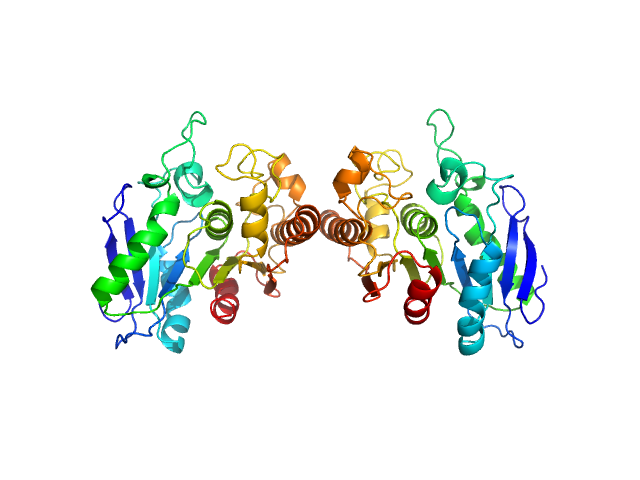
| Sample: |
Upstream of N-ras, isoform A monomer, 19 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 14
|
Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep 32(3):107930 (2020)
Hollmann NM, Jagtap PKA, Masiewicz P, Guitart T, Simon B, Provaznik J, Stein F, Haberkant P, Sweetapple LJ, Villacorta L, Mooijman D, Benes V, Savitski MM, Gebauer F, Hennig J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly(Aspartic acid) hydrolase-1 dimer, 65 kDa Sphingomonas sp. KT-1 protein
|
| Buffer: |
20 mM Tris pH 7.0, 100 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 27
|
Structural Characterization of Sphingomonas
sp. KT-1 PahZ1-Catalyzed Biodegradation of Thermally Synthesized Poly(aspartic acid)
ACS Sustainable Chemistry & Engineering (2020)
Brambley C, Bolay A, Salvo H, Jansch A, Yared T, Miller J, Wallen J, Weiland M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Pentafunctional AROM polypeptide dimer, 345 kDa Chaetomium thermophilum protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl. 2 mM TCEP, 1% sucrose, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Nov 22
|
Architecture and functional dynamics of the pentafunctional AROM complex
Nature Chemical Biology (2020)
Arora Verasztó H, Logotheti M, Albrecht R, Leitner A, Zhu H, Hartmann M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
623 |
nm3 |
|
|