|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl, pH: 6.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 13
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
346 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl, pH: 6.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jul 13
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
429 |
nm3 |
|
|
|
|
|
|
|
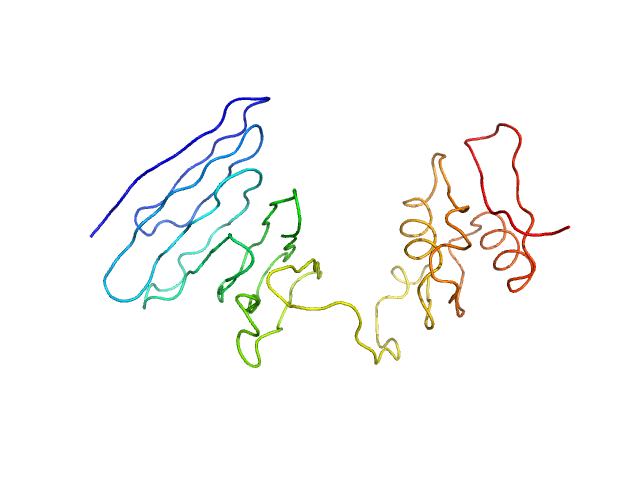
| Sample: |
Severe fever with thrombocytopenia syndrome virus L protein - RNA-dependent RNA polymerase monomer, 238 kDa SFTS virus AH12 protein
|
| Buffer: |
50 mM HEPES(NaOH) pH 7.0, 500 mM NaCl, 5% (w/v) glycerol, and 2 mM dithiothreitol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 13
|
Structural and functional characterization of the Severe fever with thrombocytopenia syndrome virus L protein
(2020)
Vogel D, Thorkelsson S, Quemin E, Meier K, Kouba T, Gogrefe N, Busch C, Reindl S, Günther S, Cusack S, Grünewald K, Rosenthal M
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
483 |
nm3 |
|
|
|
|
|
|
|
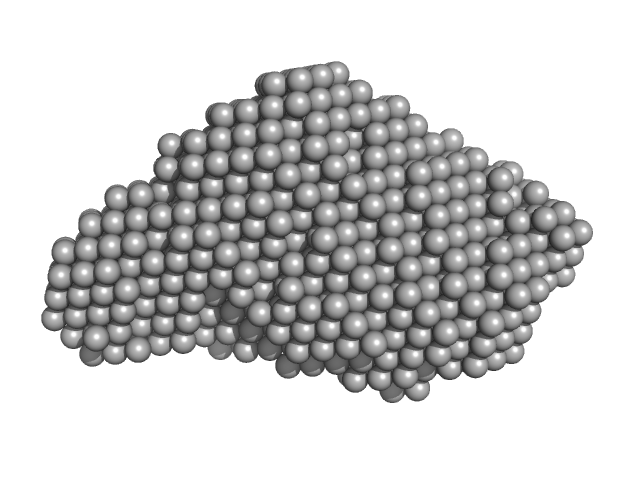
| Sample: |
2,4-dichlorophenol 6-monooxygenase hexamer, 399 kDa Streptomyces sp. SCSIO … protein
Flavin adenine dinucleotide hexamer, 5 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5 mM DTT, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Oct 22
|
Structural analyses of the group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J Biol Chem (2020)
Manenda MS, Picard MÈ, Zhang L, Cyr N, Zhu X, Barma J, Pascal JM, Couture M, Zhang C, Shi R
|
| RgGuinier |
4.8 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
624 |
nm3 |
|
|
|
|
|
|
|
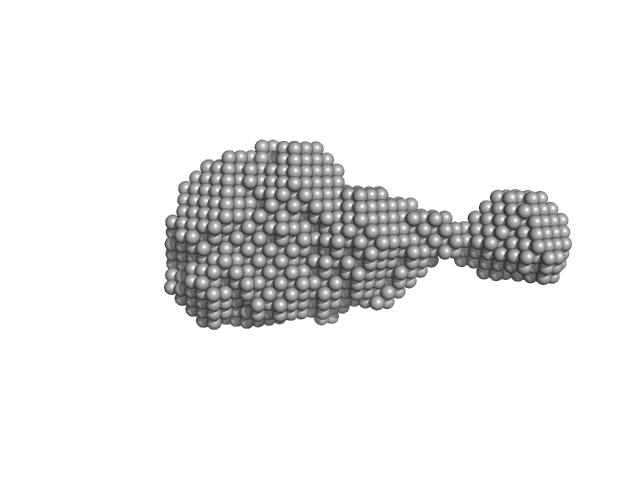
| Sample: |
PupR protein monomer, 24 kDa Pseudomonas putida protein
|
| Buffer: |
25 mM HEPES 400 mM LiCl 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Mar 16
|
Structural basis of cell surface signaling by a conserved sigma regulator in Gram-negative bacteria.
J Biol Chem (2020)
Jensen JL, Jernberg BD, Sinha S, Colbert CL
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
PupR protein monomer, 24 kDa Pseudomonas putida protein
Ferric-pseudobactin BN7/BN8 receptor monomer, 8 kDa Pseudomonas putida protein
|
| Buffer: |
25 mM HEPES 400 mM LiCl 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Mar 16
|
Structural basis of cell surface signaling by a conserved sigma regulator in Gram-negative bacteria.
J Biol Chem (2020)
Jensen JL, Jernberg BD, Sinha S, Colbert CL
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
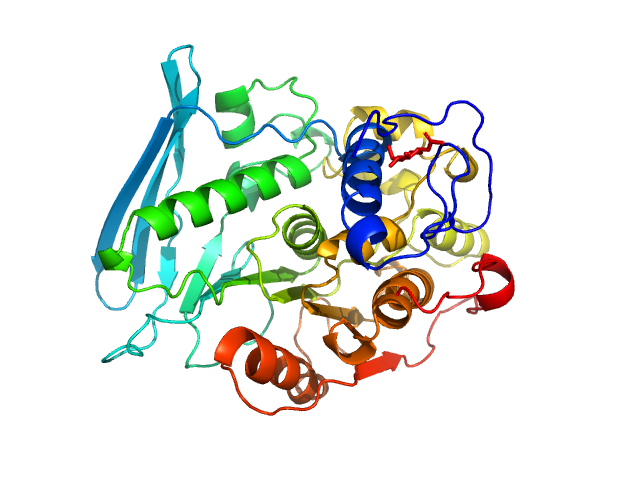
| Sample: |
4-O-methyl-glucuronoyl methylesterase (Glucuronoyl esterase) monomer, 51 kDa Cerrena unicolor protein
|
| Buffer: |
20 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 10
|
The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun 11(1):1026 (2020)
Ernst HA, Mosbech C, Langkilde AE, Westh P, Meyer AS, Agger JW, Larsen S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
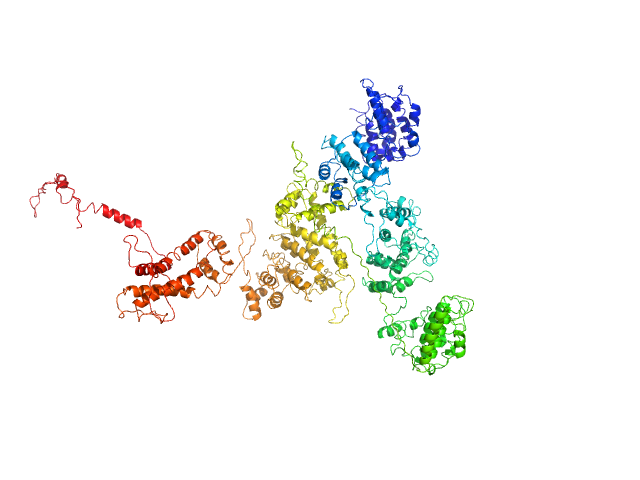
| Sample: |
4-O-methyl-glucuronoyl methylesterase (Glucuronoyl esterase, truncated) monomer, 43 kDa Cerrena unicolor protein
|
| Buffer: |
20 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 10
|
The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun 11(1):1026 (2020)
Ernst HA, Mosbech C, Langkilde AE, Westh P, Meyer AS, Agger JW, Larsen S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Macrophage mannose receptor 1 dimer, 315 kDa Mouse myeloma cell … protein
|
| Buffer: |
50mM Hepes, 100mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 15
|
Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses.
Sci Transl Med 12(530) (2020)
Jaynes JM, Sable R, Ronzetti M, Bautista W, Knotts Z, Abisoye-Ogunniyan A, Li D, Calvo R, Dashnyam M, Singh A, Guerin T, White J, Ravichandran S, Kumar P, Talsania K, Chen V, Ghebremedhin A, Karanam B, Bin Salam A, Amin R, Odzorig T, Aiken T, Nguyen V, Bian Y, Zarif JC, de Groot AE, Mehta M, Fan L, Hu X, Simeonov A, Pate N, Abu-Asab M, Ferrer M, Southall N, Ock CY, Zhao Y, Lopez H, Kozlov S, de Val N, Yates CC, Baljinnyam B, Marugan J, Rudloff U
|
| RgGuinier |
7.9 |
nm |
| Dmax |
30.1 |
nm |
| VolumePorod |
584 |
nm3 |
|
|
|
|
|
|

|
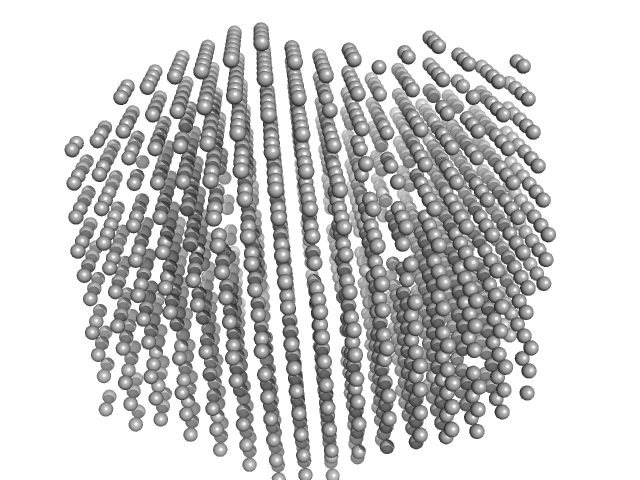
| Sample: |
Lysozyme amyloid fibril, 14 kDa Gallus gallus protein
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 5
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
30.2 |
nm |
| Dmax |
120.0 |
nm |
|
|