|
|
|
|
|
| Sample: |
Fe3O4 nanoparticles; radius 5.6 nm (AFM based) monomer, 1 kDa
|
| Buffer: |
water, HCLO4, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 5
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
11.0 |
nm |
| Dmax |
20.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; radius 5.6 nm (AFM based) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 8
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
29.4 |
nm |
| Dmax |
80.0 |
nm |
|
|
|
|
|
|
|
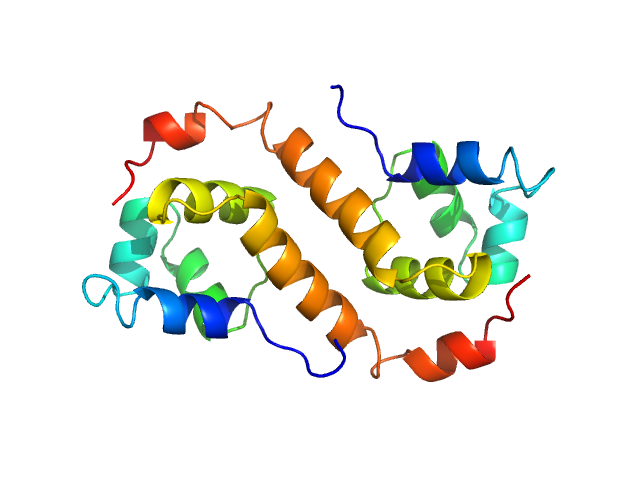
| Sample: |
Uncharacterized protein dimer, 22 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 5% (v/v) glycerol, and 1 mM PMSF, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Dec 21
|
Structural Insights Into the Transcriptional Regulation of HigBA Toxin–Antitoxin System by Antitoxin HigA in Pseudomonas aeruginosa
Frontiers in Microbiology 10 (2020)
Liu Y, Gao Z, Liu G, Geng Z, Dong Y, Zhang H
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
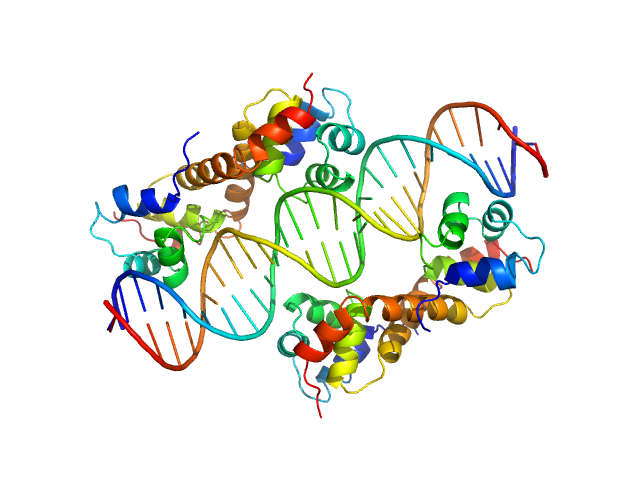
| Sample: |
Uncharacterized protein dimer, 22 kDa Pseudomonas aeruginosa protein
DNA Duplex dimer, 20 kDa DNA
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 5% (v/v) glycerol, and 1 mM PMSF, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Sep 19
|
Structural Insights Into the Transcriptional Regulation of HigBA Toxin–Antitoxin System by Antitoxin HigA in Pseudomonas aeruginosa
Frontiers in Microbiology 10 (2020)
Liu Y, Gao Z, Liu G, Geng Z, Dong Y, Zhang H
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|

|
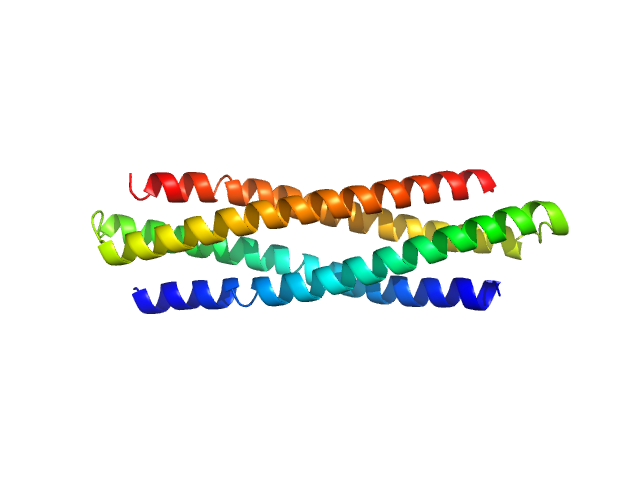
| Sample: |
De novo protein WA20 dimer, 25 kDa de novo protein protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 200 mM ArgHCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-6A, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2016 Feb 26
|
Hyperstable De Novo Protein with a Dimeric Bisecting Topology.
ACS Synth Biol (2020)
Kimura N, Mochizuki K, Umezawa K, Hecht MH, Arai R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Hyperstable de novo protein Super WA20 dimer, 25 kDa de novo protein protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 200 mM ArgHCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-6A, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2016 Feb 26
|
Hyperstable De Novo Protein with a Dimeric Bisecting Topology.
ACS Synth Biol (2020)
Kimura N, Mochizuki K, Umezawa K, Hecht MH, Arai R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGS5_fit1_model1.png)
|
| Sample: |
MvaT(mutant) dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20 mM Bis-Tris 50 mM KCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 11
|
Structural basis for osmotic regulation of the DNA binding properties of H-NS proteins.
Nucleic Acids Res (2020)
Qin L, Bdira FB, Sterckx YGJ, Volkov AN, Vreede J, Giachin G, van Schaik P, Ubbink M, Dame RT
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGT5_fit1_model1.png)
|
| Sample: |
MvaT(mutant) dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20 mM Bis-Tris 300 mM KCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 11
|
Structural basis for osmotic regulation of the DNA binding properties of H-NS proteins.
Nucleic Acids Res (2020)
Qin L, Bdira FB, Sterckx YGJ, Volkov AN, Vreede J, Giachin G, van Schaik P, Ubbink M, Dame RT
|
| RgGuinier |
3.8 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
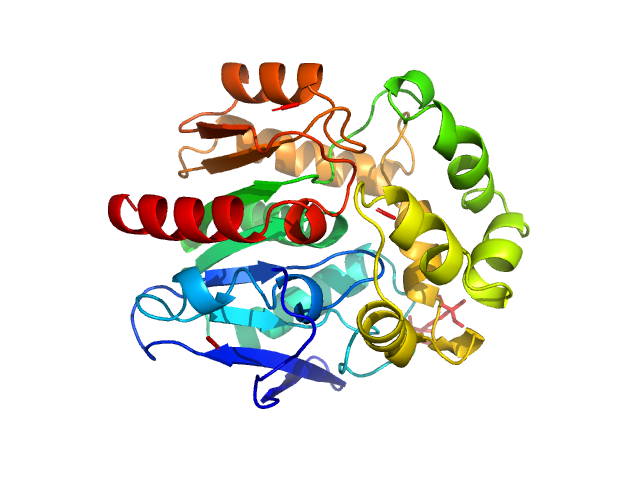
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 56 kDa Rattus norvegicus protein
Guanine nucleotide-binding protein G(i) subunit alpha-1 monomer, 38 kDa Rattus norvegicus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 30
|
Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to Gαi1
Nature Communications 11(1) (2020)
McClelland L, Zhang K, Mou T, Johnston J, Yates-Hansen C, Li S, Thomas C, Doukov T, Triest S, Wohlkonig A, Tall G, Steyaert J, Chiu W, Sprang S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Haloalkane dehalogenase variant DhaA115 -monomeric fraction monomer, 34 kDa Rhodococcus rhodochrous protein
|
| Buffer: |
50 mM potassium phosphate buffer (41 mM K₂HPO₄, 9mM KH₂PO₄), pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2019 Aug 22
|
Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst
Chemical Science 11(41):11162-11178 (2020)
Markova K, Chmelova K, Marques S, Carpentier P, Bednar D, Damborsky J, Marek M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
41 |
nm3 |
|
|