|
|
|
|
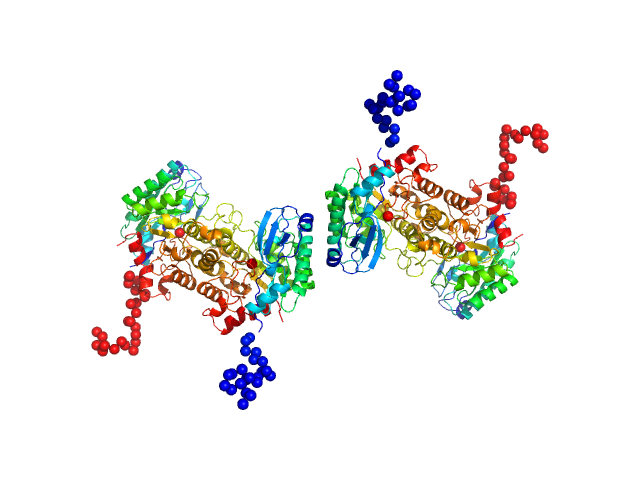
|
| Sample: |
2-amino-3-carboxymuconate 6-semialdehyde decarboxylase tetramer, 159 kDa Pseudomonas fluorescens protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Jul 15
|
Quaternary structure of α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase (ACMSD) controls its activity.
J Biol Chem 294(30):11609-11621 (2019)
Yang Y, Davis I, Matsui T, Rubalcava I, Liu A
|
| RgGuinier |
5.2 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
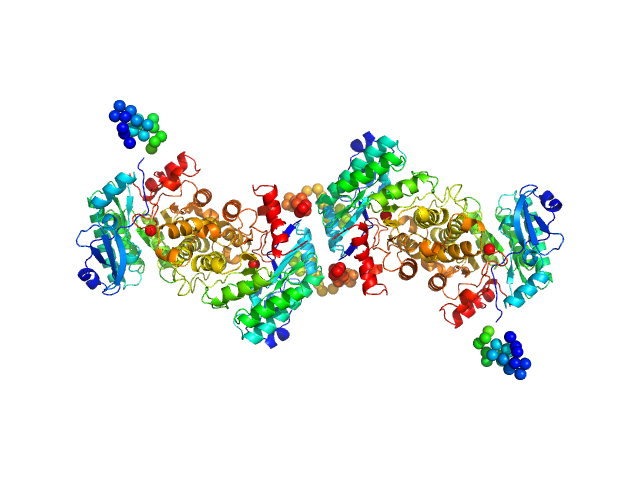
|
| Sample: |
2-amino-3-carboxymuconate 6-semialdehyde decarboxylase tetramer, 159 kDa Pseudomonas fluorescens protein
|
| Buffer: |
25 mM HEPES, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Jan 10
|
Quaternary structure of α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase (ACMSD) controls its activity.
J Biol Chem 294(30):11609-11621 (2019)
Yang Y, Davis I, Matsui T, Rubalcava I, Liu A
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
195 |
nm3 |
|
|
|
|
|
|
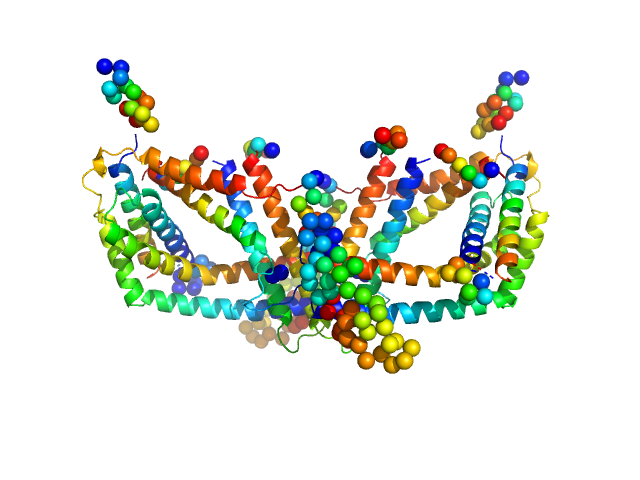
|
| Sample: |
Polyphosphate-targeting protein A dimer, 79 kDa Streptomyces chartreusis protein
|
| Buffer: |
20 mM Tris-HCl 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 24
|
Structural and biochemical analysis of a phosin from Streptomyces chartreusis reveals a combined polyphosphate- and metal-binding fold.
FEBS Lett (2019)
Werten S, Rustmeier NH, Gemmer M, Virolle MJ, Hinrichs W
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
|
|
|
|
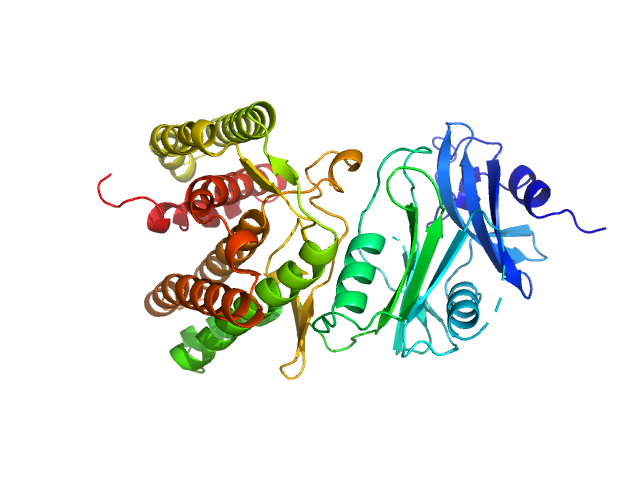
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
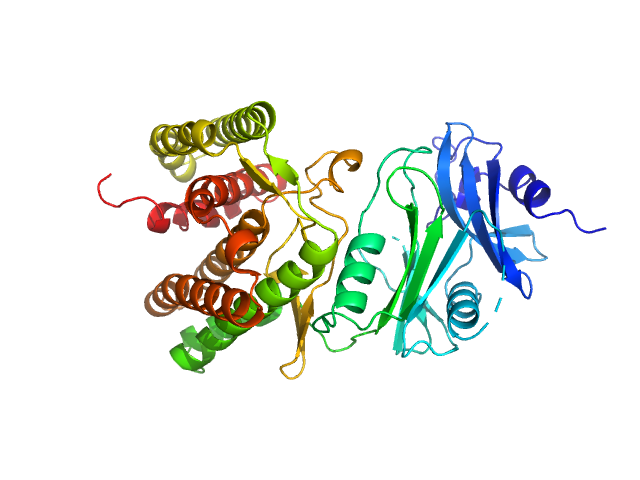
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 0.2 M D-glucosamine, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 20
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
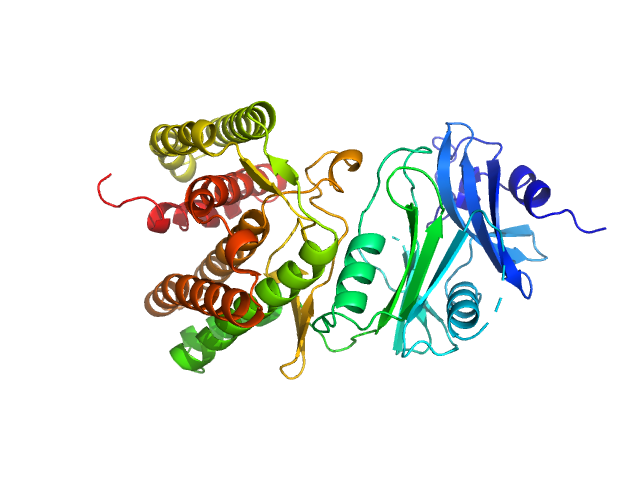
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
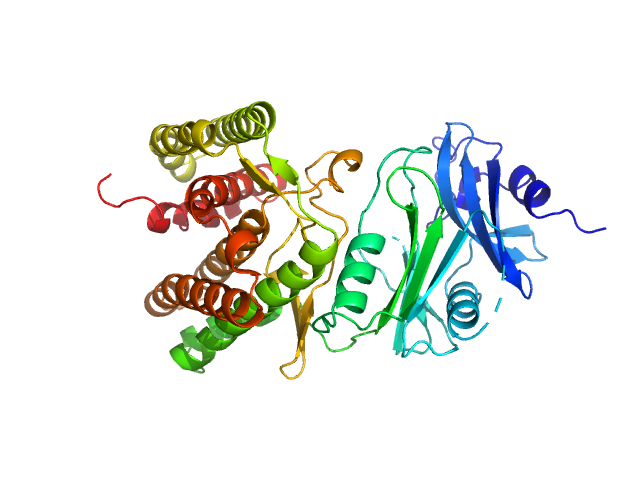
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 0.2 M D-glucosamine, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 20
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
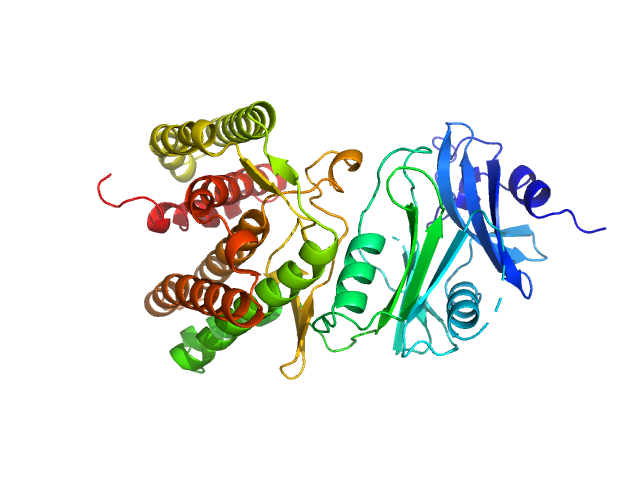
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM D-glucose, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Apr 13
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM D-glucose, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 51 kDa Rattus norvegicus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Apr 24
|
Structure, Function, and Dynamics of the Gα Binding Domain of Ric-8A.
Structure (2019)
Zeng B, Mou TC, Doukov TI, Steiner A, Yu W, Papasergi-Scott M, Tall GG, Hagn F, Sprang SR
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
70 |
nm3 |
|
|