|
|
|
|
|
| Sample: |
Truncated P5abc subdomain from tetrahymena ribozyme monomer, 18 kDa RNA
|
| Buffer: |
0.5mM MgCl2 20mM KCl 10mM KMOPS 20uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Jun 17
|
Revealing the distinct folding phases of an RNA three-helix junction.
Nucleic Acids Res 46(14):7354-7365 (2018)
Plumridge A, Katz AM, Calvey GD, Elber R, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Truncated P5abc subdomain from tetrahymena ribozyme monomer, 18 kDa RNA
|
| Buffer: |
1mM MgCl2 20mM KCl 10mM KMOPS 20uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Jun 17
|
Revealing the distinct folding phases of an RNA three-helix junction.
Nucleic Acids Res 46(14):7354-7365 (2018)
Plumridge A, Katz AM, Calvey GD, Elber R, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
|
|
|
|
|
|

|
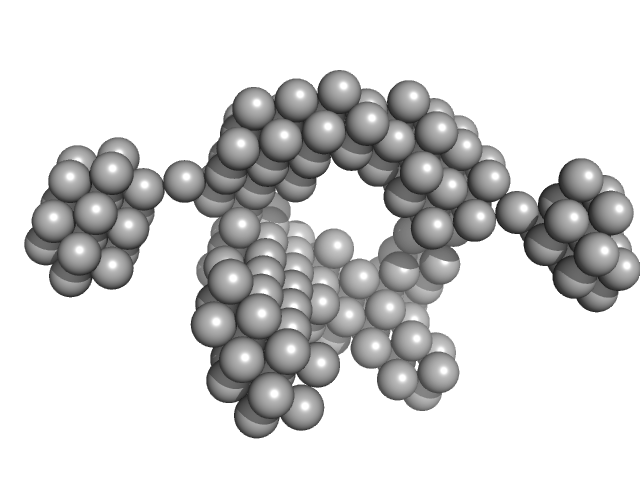
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M KCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M NaCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
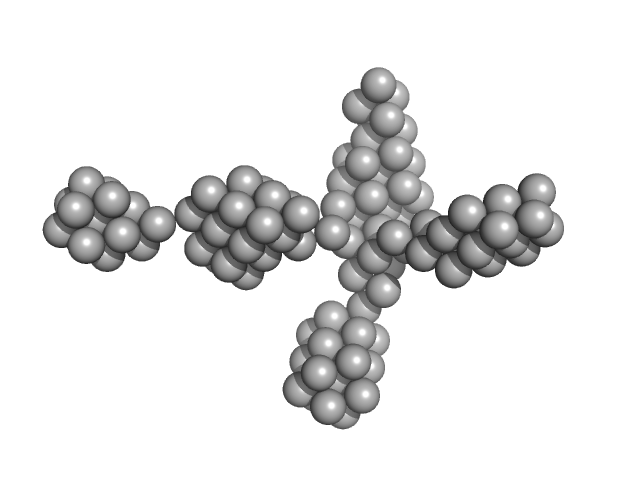
|
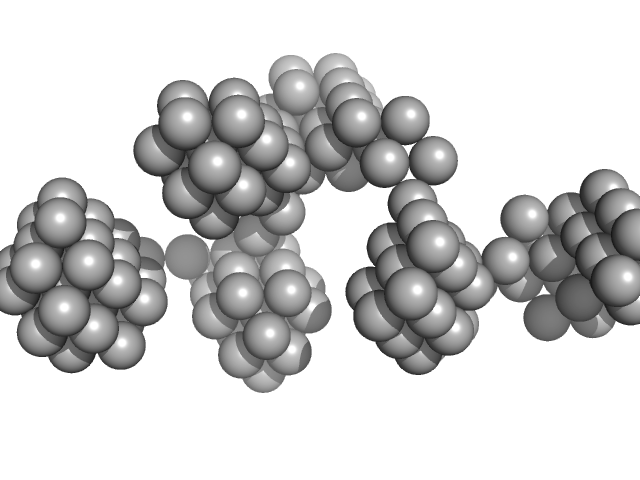
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M KBr, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
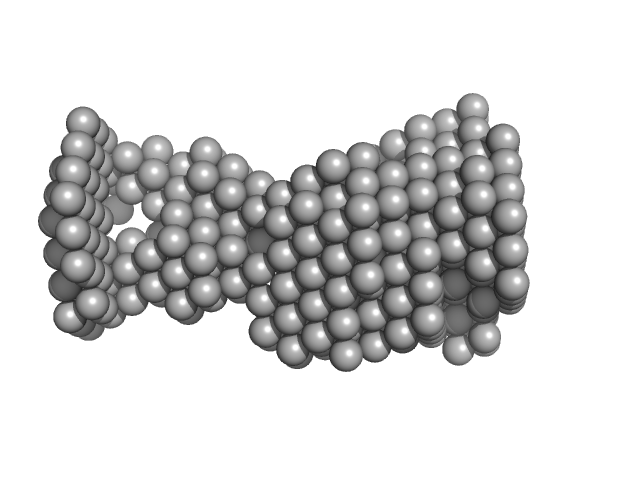
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M NaBr, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 26
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M RbCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin Protease monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 6
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 6
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
152 |
nm3 |
|
|