|
|
|
|
|
| Sample: |
Type III secretion protein HrpG, 19 kDa Erwinia amylovora protein
Type III secretion protein HrpV, 13 kDa Erwinia amylovora protein
Hypersensitivity response secretion protein HrpJ, 42 kDa Erwinia amylovora protein
|
| Buffer: |
20mM Tris-Cl, pH 8.0, 100mM NaCl, 2mM DTT, 0.5mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 3
|
Migration of Type III Secretion System Transcriptional Regulators Links Gene Expression to Secretion.
MBio 9(4) (2018)
Charova SN, Gazi AD, Mylonas E, Pozidis C, Sabarit B, Anagnostou D, Psatha K, Aivaliotis M, Beuzon CR, Panopoulos NJ, Kokkinidis M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Fusion protein of LSm and MyoX-coil, 1066 kDa Artificial protein protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-6A, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Nov 11
|
Design of Hollow Protein Nanoparticles with Modifiable Interior and Exterior Surfaces.
Angew Chem Int Ed Engl (2018)
Kawakami N, Kondo H, Matsuzawa Y, Hayasaka K, Nasu E, Sasahara K, Arai R, Miyamoto K
|
| RgGuinier |
9.4 |
nm |
| Dmax |
23.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
|
|
|
|
|
| Sample: |
Ovalbumin (common quail) monomer, 42 kDa Coturnix coturnix protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
|
|
|
|
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
|
|
|
|

|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
0.16 M NaOH, pH: 13.2 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Feb 21
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
|
|
|
|

|
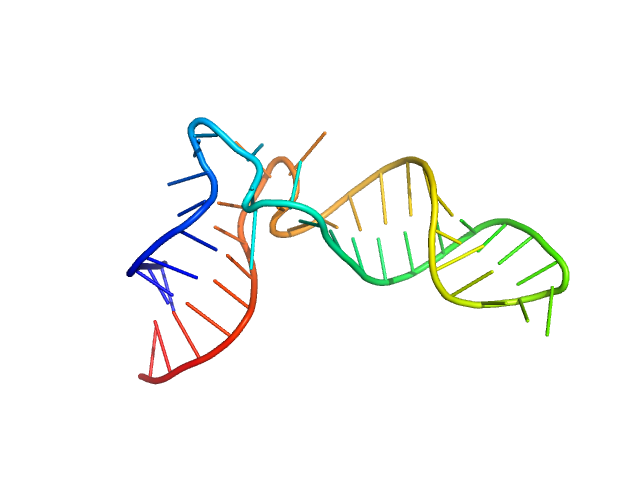
| Sample: |
Arginyl transfer RNA monomer, 13 kDa Romanomermis culicivorax RNA
|
| Buffer: |
50 mM HEPES-NaOH 10 mM MgCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Dec 16
|
Small but large enough: structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax.
Nucleic Acids Res (2018)
Jühling T, Duchardt-Ferner E, Bonin S, Wöhnert J, Pütz J, Florentz C, Betat H, Sauter C, Mörl M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoleucyl transfer RNA monomer, 15 kDa Romanomermis culicivorax RNA
|
| Buffer: |
50 mM HEPES-NaOH 10 mM MgCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Dec 16
|
Small but large enough: structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax.
Nucleic Acids Res (2018)
Jühling T, Duchardt-Ferner E, Bonin S, Wöhnert J, Pütz J, Florentz C, Betat H, Sauter C, Mörl M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase from Clostridium botulinum tetramer, 126 kDa Clostridium botulinum protein
|
| Buffer: |
20mM Tris, 150mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2010 Nov 26
|
Substrate Locking Promotes Dimer-Dimer Docking of an Enzyme Antibiotic Target.
Structure 26(7):948-959.e5 (2018)
Atkinson SC, Dogovski C, Wood K, Griffin MDW, Gorman MA, Hor L, Reboul CF, Buckle AM, Wuttke J, Parker MW, Dobson RCJ, Perugini MA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase from Clostridium botulinum tetramer, 126 kDa Clostridium botulinum protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 5mM sodium pyruvate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2010 Nov 26
|
Substrate Locking Promotes Dimer-Dimer Docking of an Enzyme Antibiotic Target.
Structure 26(7):948-959.e5 (2018)
Atkinson SC, Dogovski C, Wood K, Griffin MDW, Gorman MA, Hor L, Reboul CF, Buckle AM, Wuttke J, Parker MW, Dobson RCJ, Perugini MA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
165 |
nm3 |
|
|