|
|
|
|
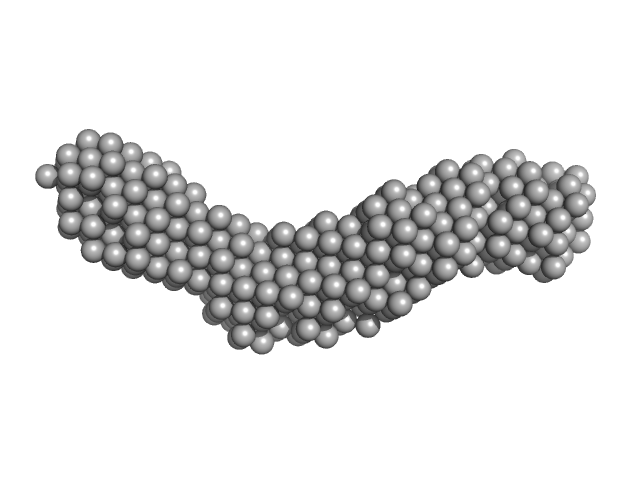
|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
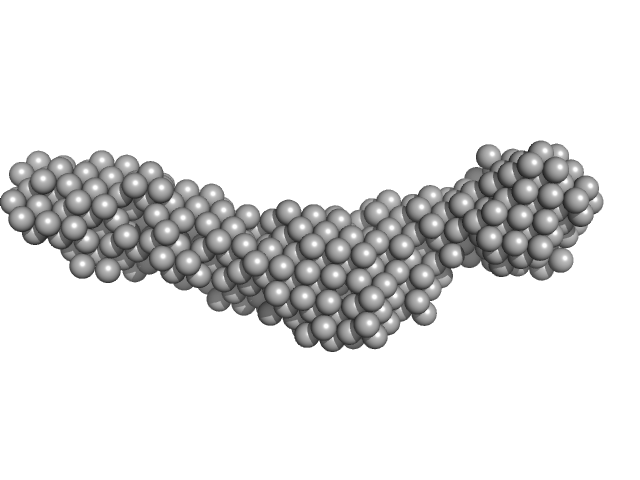
|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
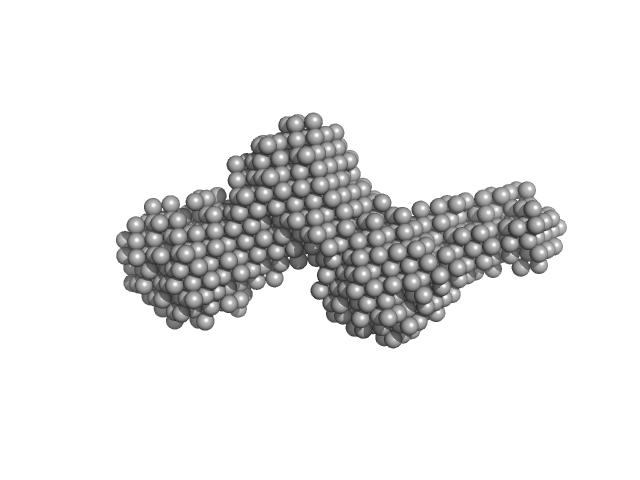
|
| Sample: |
Collagenase ColG segement s2s3as3b monomer, 37 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColG segement s2s3as3b monomer, 37 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
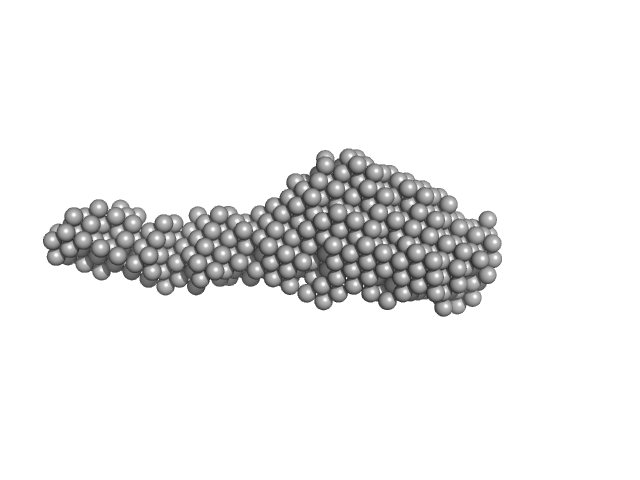
|
| Sample: |
Collagenase ColG segement s2s3as3b monomer, 37 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
4.3 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColG segement s2s3as3b monomer, 37 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
N-terminus of disulfide interchange protein DsbD monomer, 14 kDa Neisseria meningitidis protein
|
| Buffer: |
25mM HEPES 150mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Production, biophysical characterization and initial crystallization studies of the N- and C-terminal domains of DsbD, an essential enzyme in Neisseria meningitidis.
Acta Crystallogr F Struct Biol Commun 74(Pt 1):31-38 (2018)
Smith RP, Whitten AE, Paxman JJ, Kahler CM, Scanlon MJ, Heras B
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein) monomer, 21 kDa Wolbachia endosymbiont of … protein
|
| Buffer: |
25mM HEPES 150mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Production, biophysical characterization and initial crystallization studies of the N- and C-terminal domains of DsbD, an essential enzyme in Neisseria meningitidis.
Acta Crystallogr F Struct Biol Commun 74(Pt 1):31-38 (2018)
Smith RP, Whitten AE, Paxman JJ, Kahler CM, Scanlon MJ, Heras B
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|