|
|
|
|
|
| Sample: |
EspG3 chaperone from Mycobacterium marinum M monomer, 32 kDa Mycobacterium marinum M protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
EspG1 from Mycobacterium marinum monomer, 30 kDa Mycobacterium marinum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
EspG3 chaperone from Mycobacterium smegmatis monomer, 32 kDa Mycobacterium smegmatis protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
EspG3 chaperone from Mycobacterium smegmatis monomer, 32 kDa Mycobacterium smegmatis protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.2 |
nm |
|
|
|
|
|
|
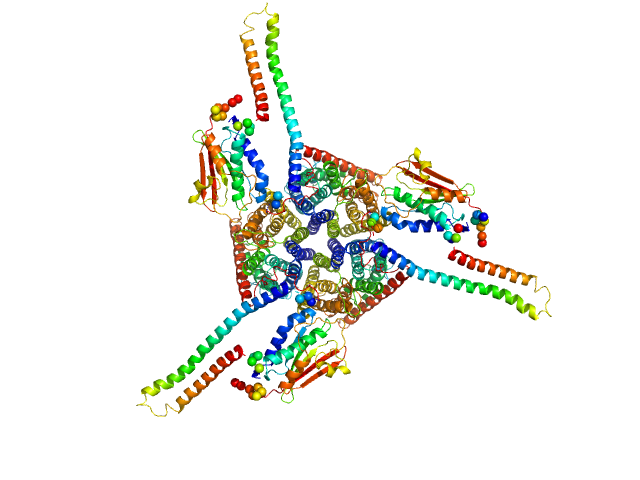
|
| Sample: |
Ammonium sensor/transducer trimer, 227 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, 0.09%(w/v) CYMAL-5, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 1
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
511 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ammonium transporter histidine kinase domain monomer, 30 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ammonium transporter histidine kinase domain monomer, 30 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ammonium transporter histidine kinase domain monomer, 30 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ammonium sensor/transducer trimer, 227 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, 0.09%(w/v) CYMAL-5, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
482 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
35 |
nm3 |
|
|