|
|
|
|
|
| Sample: |
Bacteriophage phi-X174 monomer, 0 kDa protein
|
| Buffer: |
0.15 mg/mL LPS, 0.06 M NH4Cl2, 0.09 M NaCl, 0.1 M KCl, 1 mM MgS04, 1 mM CaCl2, 0.1 M Tris-HCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Structural changes of tailless bacteriophage ΦX174 during penetration of bacterial cell walls.
Proc Natl Acad Sci U S A 114(52):13708-13713 (2017)
Sun Y, Roznowski AP, Tokuda JM, Klose T, Mauney A, Pollack L, Fane BA, Rossmann MG
|
|
|
|
|
|
|
|
| Sample: |
Bacteriophage phi-X174 monomer, 0 kDa protein
|
| Buffer: |
0.15 mg/mL LPS, 0.06 M NH4Cl2, 0.09 M NaCl, 0.1 M KCl, 1 mM MgS04, 1 mM CaCl2, 0.1 M Tris-HCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Structural changes of tailless bacteriophage ΦX174 during penetration of bacterial cell walls.
Proc Natl Acad Sci U S A 114(52):13708-13713 (2017)
Sun Y, Roznowski AP, Tokuda JM, Klose T, Mauney A, Pollack L, Fane BA, Rossmann MG
|
|
|
|
|
|
|

|
| Sample: |
Nucleoplasmin core + A2 pentamer, 81 kDa Xenopus laevis protein
Histone H2A (ΔAla127) pentamer, 69 kDa Xenopus laevis protein
Histone H2B 1.1 (Ser33Thr) pentamer, 67 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Tris. 150 mM NaCl, 1 mM EDTA, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2016 Jan 7
|
Dynamic intramolecular regulation of the histone chaperone nucleoplasmin controls histone binding and release.
Nat Commun 8(1):2215 (2017)
Warren C, Matsui T, Karp JM, Onikubo T, Cahill S, Brenowitz M, Cowburn D, Girvin M, Shechter D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
402 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DHH subfamily 1 protein dimer, 70 kDa Streptococcus pneumoniae serotype … protein
|
| Buffer: |
20mM Tris, 200 mM NaCl, 5%(v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 23
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
T.maritima PDE dimer, 76 kDa Thermotoga maritima protein
|
| Buffer: |
25mM Tris 500mM NaCl 3% (v/v) glycerol 2mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 17
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.8 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
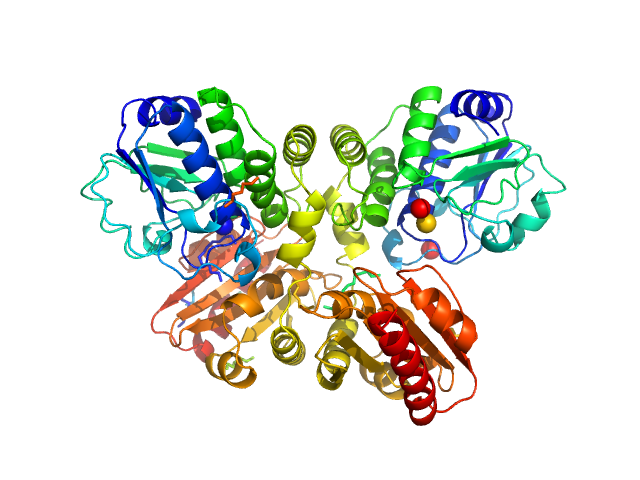
|
| Sample: |
Thermotoga maritima phosphodiesterase (wildtype, TmPDE, TM1595) dimer, 76 kDa Thermotoga maritima protein
|
| Buffer: |
20mM Tris, 200 mM NaCl, 5%(v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 23
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Telomere DNA duplex monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 50 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 May 3
|
Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it.
Nucleic Acids Res 45(21):12170-12180 (2017)
Necasová I, Janoušková E, Klumpler T, Hofr C
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
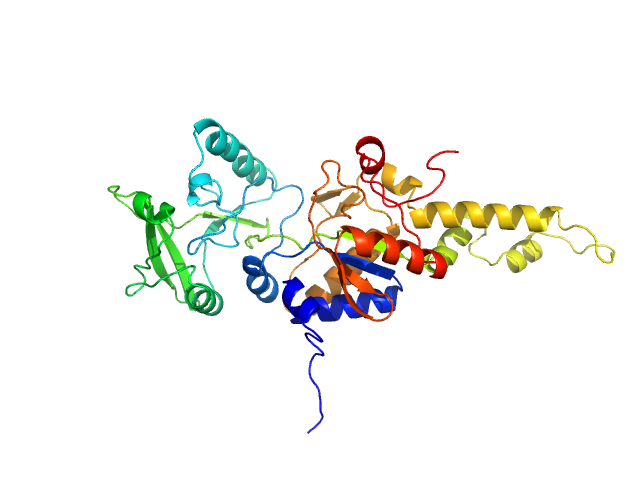
|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 39 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 4 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 19
|
Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis.
PLoS Biol 15(12):e2004486 (2017)
O'Brien DP, Durand D, Voegele A, Hourdel V, Davi M, Chamot-Rooke J, Vachette P, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Pomacea maculata perivitellin 1 dodecamer, 278 kDa Pomacea maculata protein
|
| Buffer: |
100 mM Phosphate Buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 Mar 26
|
Convergent evolution of plant and animal embryo defences by hyperstable non-digestible storage proteins.
Sci Rep 7(1):15848 (2017)
Pasquevich MY, Dreon MS, Qiu JW, Mu H, Heras H
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
537 |
nm3 |
|
|
|
|
|
|
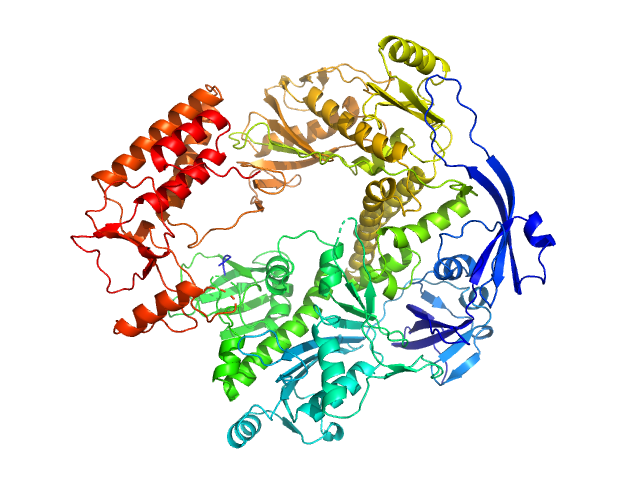
|
| Sample: |
DNA polymerase E9 monomer, 117 kDa Vaccinia virus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 4 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 27
|
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun 8(1):1455 (2017)
Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, Hart DJ, Peyrefitte CN, Burmeister WP, Iseni F
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
202 |
nm3 |
|
|