|
|
|
|

|
| Sample: |
Aureochrome1a-A´α-LOV-Jα dimer, 36 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 16
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|

|
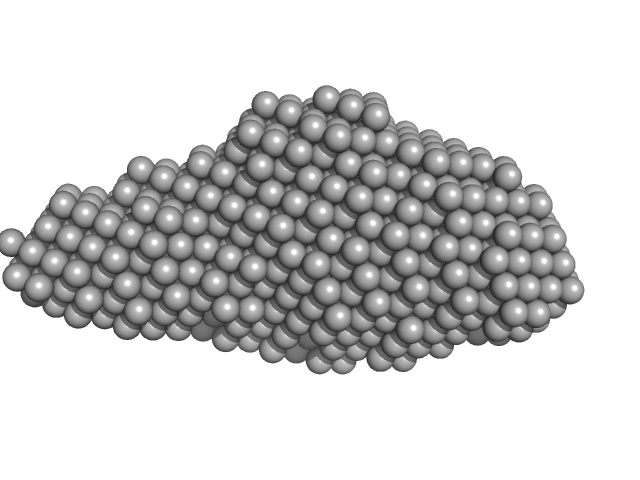
| Sample: |
Aureochrome1a-A´α-LOV-Jα dimer, 36 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 16
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aureochrome1a- A´α-LOV-Jα (Dark State) dimer, 32 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 9
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome1a- A´α-LOV-Jα (Light State) dimer, 32 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 9
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
FI10506p monomer, 23 kDa Drosophila melanogaster protein
|
| Buffer: |
20mM Tris/HCl 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 21
|
Intrinsic Disorder of the C-Terminal Domain of Drosophila Methoprene-Tolerant Protein.
PLoS One 11(9):e0162950 (2016)
Kolonko M, Ożga K, Hołubowicz R, Taube M, Kozak M, Ożyhar A, Greb-Markiewicz B
|
| RgGuinier |
5.1 |
nm |
| Dmax |
22.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Latency-associated nuclear antigen tetramer, 87 kDa Murid herpesvirus 4 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
MHV-68 TR DNA monomer, 30 kDa unidentified herpesvirus DNA
|
| Buffer: |
10 mM TRIS 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
KLBS1-2 DNA monomer, 24 kDa unidentified herpesvirus DNA
|
| Buffer: |
Tris, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
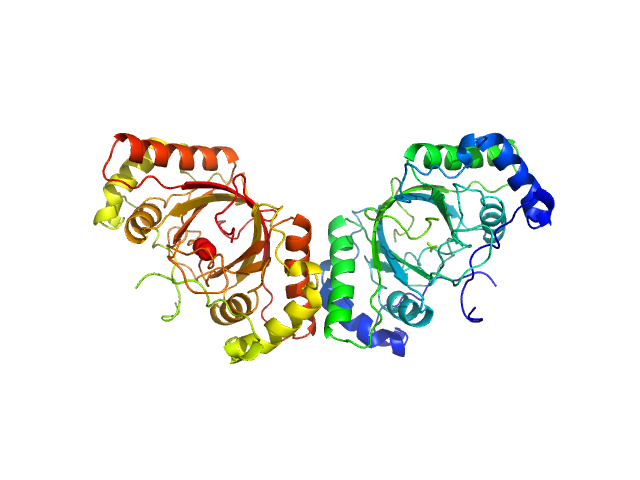
| Sample: |
ORF73 tetramer tetramer, 63 kDa Human herpesvirus 8 protein
ORF73 dimer dimer, 32 kDa Human herpesvirus 8 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
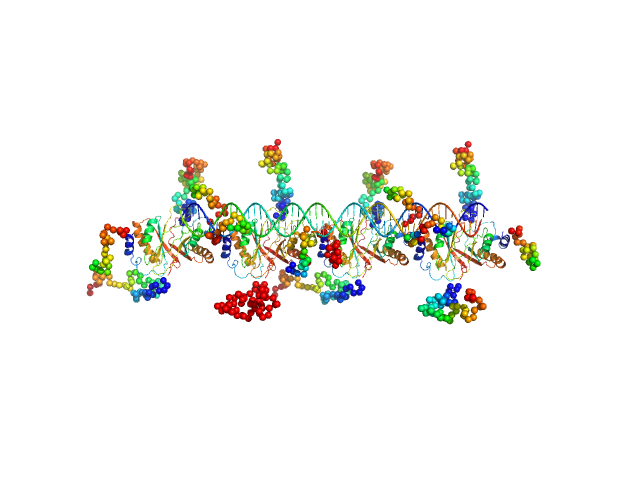
| Sample: |
MHV-68 TR DNA monomer, 30 kDa unidentified herpesvirus DNA
Latency-associated nuclear antigen octamer, 269 kDa Murid herpesvirus 4 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
5.8 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
475 |
nm3 |
|
|