|
|
|
|
|
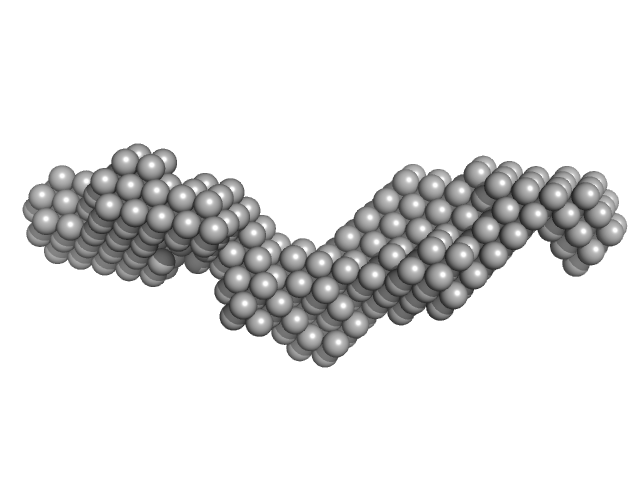
| Sample: |
Domain 3'X of hepatitis C virus monomer, 32 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.8 |
nm |
|
|
|
|
|
|

|
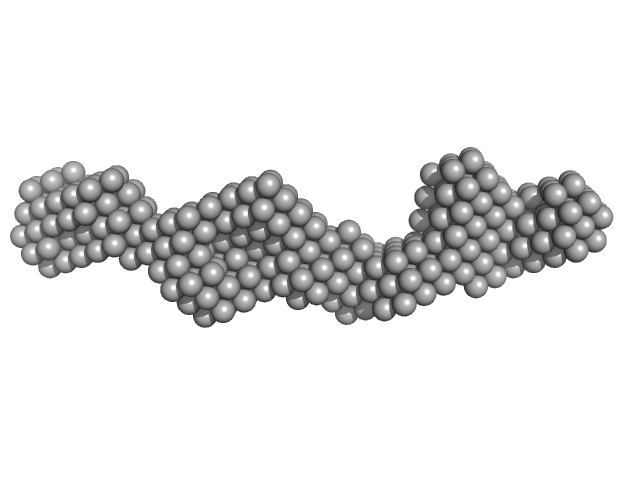
| Sample: |
Domain 3'X of hepatitis C virus dimer, 63 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA 2 mM MgCl2 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Center for Cancer Research, National Cancer Institute on 2015 Nov 20
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
6.0 |
nm |
| Dmax |
24.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
Subdomain SL2' of hepatitis C virus domain 3'X dimer, 36 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0,1mM EDTA 2mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.9 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
25 mM MOPS, 250 mM NaCl, 50 mM KCl, 2 mM TCEP, 0.1% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 9
|
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update.
Acta Crystallogr D Struct Biol 73(Pt 9):710-728 (2017)
Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
229 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Xenopus laevis protein
|
| Buffer: |
25 mM MOPS, 250 mM NaCl, 50 mM KCl, 2 mM TCEP, 0.1% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 9
|
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update.
Acta Crystallogr D Struct Biol 73(Pt 9):710-728 (2017)
Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bifunctional protein PutA dimer, 219 kDa Bdellovibrio bacteriovorus protein
|
| Buffer: |
50 mM Tris, 125 mM NaCl, 1 mM EDTA, and 1 mM tris(3-hydroxypropyl)phosphine (THP) at pH 7.5,, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Jun 8
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
287 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bifunctional protein PutA dimer, 229 kDa Desulfovibrio vulgaris protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, and 0.5 mM THP at pH 7.5., pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Jun 8
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
293 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bifunctional protein PutA dimer, 238 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, and 0.5 mM THP at pH 7.5., pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Apr 20
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
291 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Proline dehydrogenase tetramer, 430 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine, and 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 16
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
5.3 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
541 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Proline dehydrogenase tetramer, 430 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine, and 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 12
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
553 |
nm3 |
|
|