|
|
|
|
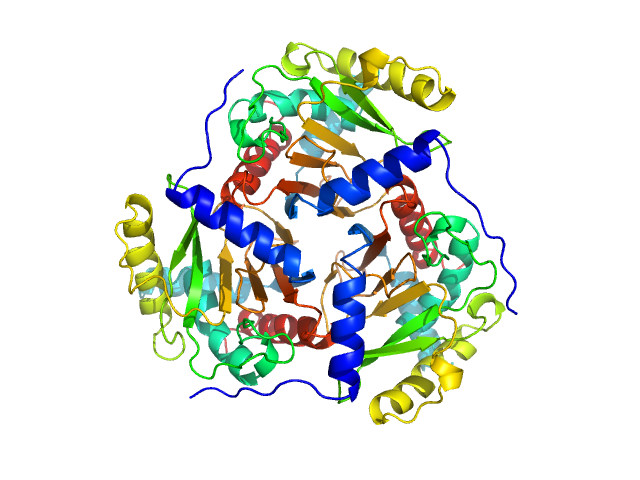
|
| Sample: |
Regulatory protein E2 trimer, 73 kDa Human papillomavirus type … protein
|
| Buffer: |
50 mM Tris ⁄ HCl, pH 8.8, 100 mM NaCl, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 27
|
Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex
Biochemical Journal 463(3):405-412 (2014)
Marrott N, Marshall J, Svergun D, Crennell S, Hough D, van den Elsen J, Danson M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
|
|
|
|
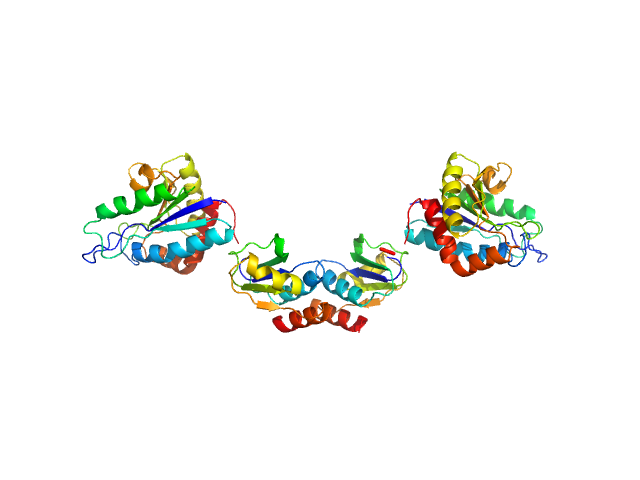
|
| Sample: |
Clostridium difficile bacteriophage 27 endolysin dimer, 64 kDa Clostridioides difficile protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 17
|
The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.
PLoS Pathog 10(7):e1004228 (2014)
Dunne M, Mertens HD, Garefalaki V, Jeffries CM, Thompson A, Lemke EA, Svergun DI, Mayer MJ, Narbad A, Meijers R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
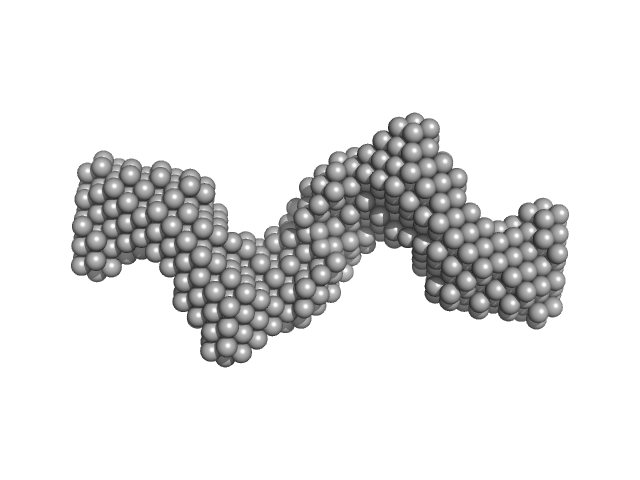
|
| Sample: |
Clostridium difficile bacteriophage 27 endolysin C238R mutant dimer, 64 kDa Clostridioides difficile protein
|
| Buffer: |
20 mM HEPES 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 21
|
The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.
PLoS Pathog 10(7):e1004228 (2014)
Dunne M, Mertens HD, Garefalaki V, Jeffries CM, Thompson A, Lemke EA, Svergun DI, Mayer MJ, Narbad A, Meijers R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Chitinase 60 monomer, 61 kDa Moritella marina protein
|
| Buffer: |
20 mM Tris 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 18
|
Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution.
Acta Crystallogr D Biol Crystallogr 70(Pt 3):676-84 (2014)
Malecki PH, Vorgias CE, Petoukhov MV, Svergun DI, Rypniewski W
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
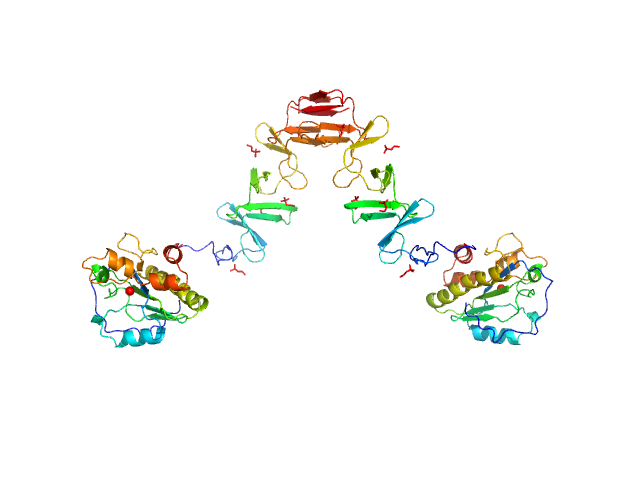
|
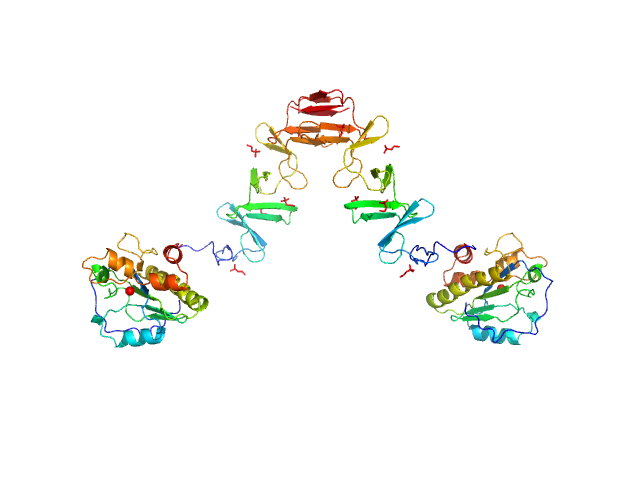
| Sample: |
Lytic Amidase with choline dimer, 75 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
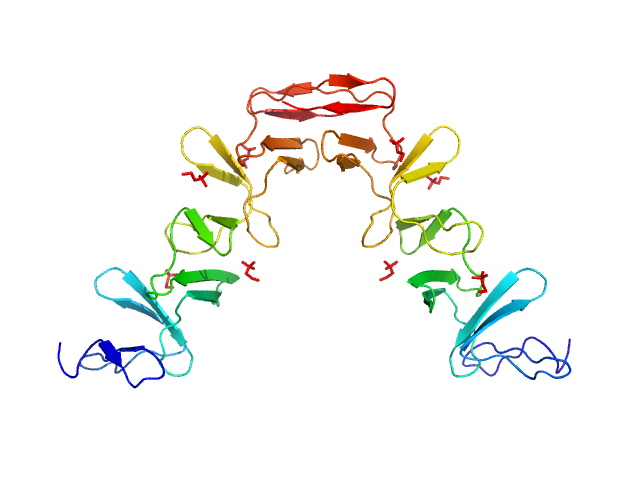
| Sample: |
Prophage Lytic Amidase with choline dimer, 73 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
5.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
|
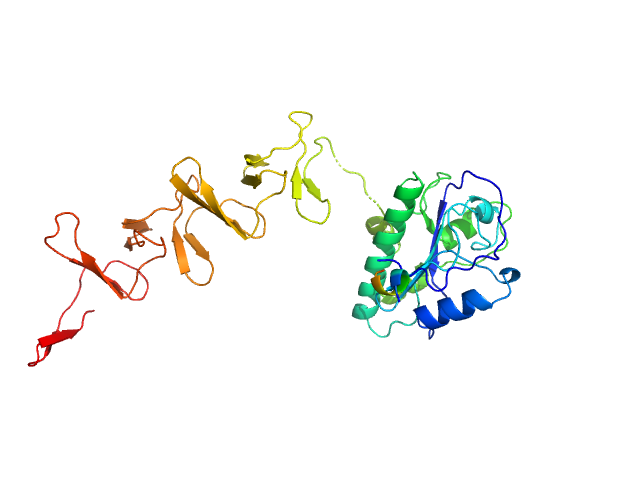
| Sample: |
Lytic Amidase without choline monomer, 37 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 3
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
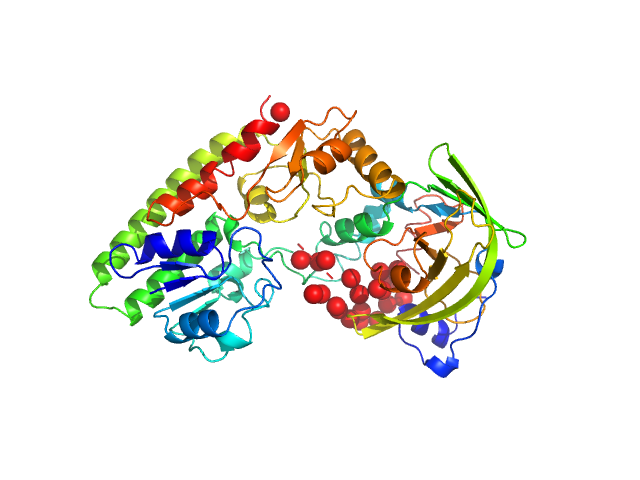
| Sample: |
Lytic Amidase choline-binding domain dimer, 17 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 11
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Functional binding region (187-385) of the pneumococcal serine-rich repeat protein monomer, 22 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM sodium citrate 250 mM NaCl 2.5 % Glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
The basic keratin 10-binding domain of the virulence-associated pneumococcal serine-rich protein PsrP adopts a novel MSCRAMM fold.
Open Biol 4:130090 (2014)
Schulte T, Löfling J, Mikaelsson C, Kikhney A, Hentrich K, Diamante A, Ebel C, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
High-affinity zinc transporter periplasmic component monomer, 33 kDa Salmonella enterica subsp. … protein
Zinc/cadmium-binding protein monomer, 23 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 May 9
|
The Salmonella enterica ZinT structure, zinc affinity and interaction with the high-affinity uptake protein ZnuA provide insight into the management of periplasmic zinc.
Biochim Biophys Acta 1840(1):535-44 (2014)
Ilari A, Alaleona F, Tria G, Petrarca P, Battistoni A, Zamparelli C, Verzili D, Falconi M, Chiancone E
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|