|
|
|
|
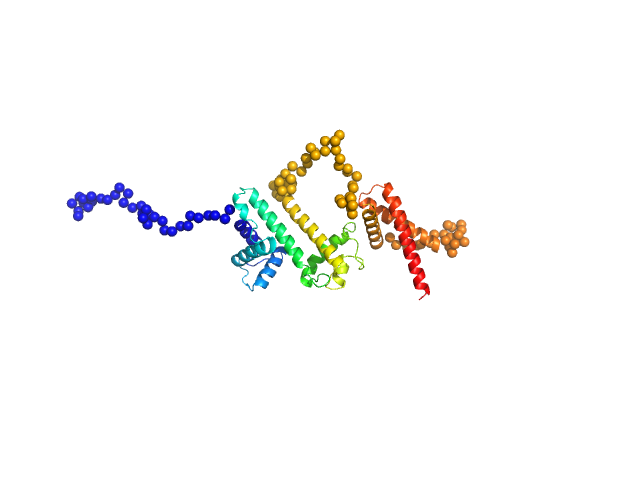
|
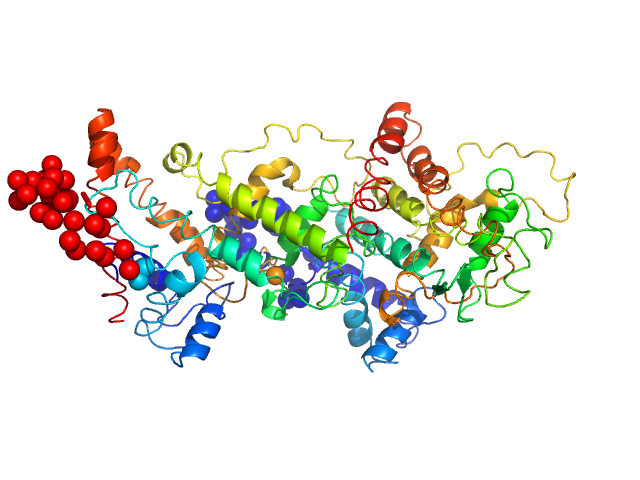
| Sample: |
Harpin Z2 monomer, 37 kDa Pseudomonas syringae strain … protein
|
| Buffer: |
10 mM MES, 100 mM NaF, pH: 6.2 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Centre for Cellular and Molecular Biology on 2023 May 1
|
Solution conformation of HrpZ2 protein from Pseudomonas syringae
Arpita Goswami
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Harpin Z2 monomer, 37 kDa Pseudomonas syringae strain … protein
|
| Buffer: |
10 mM MES, 100 mM NaF, pH: 6.2 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Centre for Cellular and Molecular Biology on 2023 May 1
|
Solution conformation of HrpZ2 protein from Pseudomonas syringae
Arpita Goswami
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphocholine hydrolase Lem3 monomer, 63 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Sep 20
|
The structural analysis of dephosphocholinase Legionella pneumophila Lem3
Wenhua Zhang
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|

|
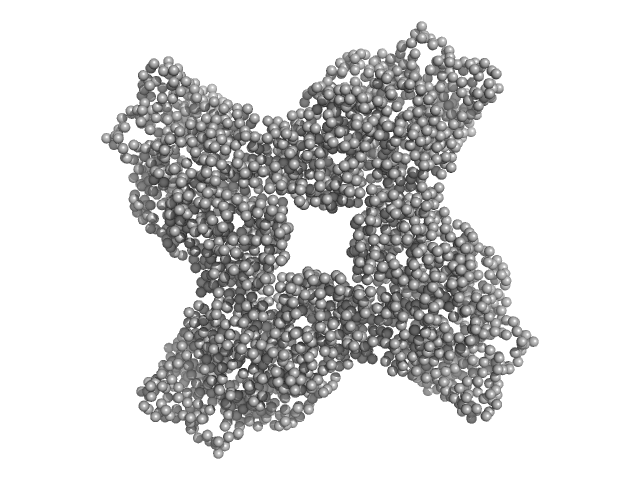
| Sample: |
Uncharacterized protein SAUSA300_1119 dimer, 125 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM KCl, 1 mM TCEP, 5% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 5
|
Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase.
J Biol Chem 300(12):107920 (2024)
Myers MJ, Xu Z, Ryan BJ, DeMars ZR, Ridder MJ, Johnson DK, Krute CN, Flynn TS, Kashipathy MM, Battaile KP, Schnicker N, Lovell S, Freudenthal BD, Bose JL
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
|
|
|
|
|
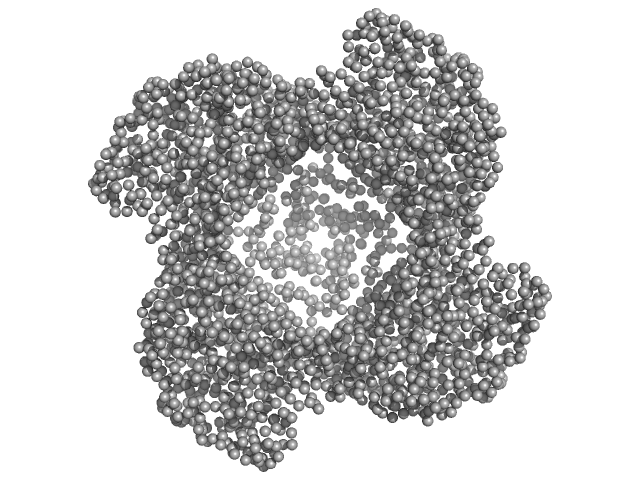
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
439 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
435 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
446 |
nm3 |
|
|
|
|
|
|
|
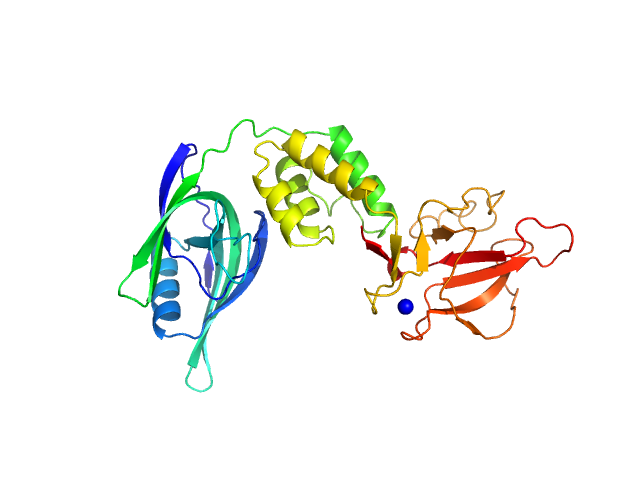
| Sample: |
Cereblon-midi monomer, 37 kDa protein
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
G-quadrupex monomer, 6 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 15
|
G-quadruplex from HBV genome
Trushar Patel
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
G-quadruplex mutant monomer, 6 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 15
|
G-quadruplex from HBV genome
Trushar Patel
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
11 |
nm3 |
|
|