|
|
|
|
|
| Sample: |
SAM riboswitch element A aptamer domain (RNA) monomer, 39 kDa Listeria monocytogenes RNA
|
| Buffer: |
50 mM potassium phosphate, 135 mM NaCl, 5 mM MgCl2, pH: 6.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 9
|
Functional Validation of SAM Riboswitch Element A from Listeria monocytogenes.
Biochemistry (2024)
Hall I, Zablock K, Sobetski R, Weidmann CA, Keane SC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SAM riboswitch element A aptamer domain (RNA) monomer, 39 kDa Listeria monocytogenes RNA
S-adenosyl-L-methionine monomer, 0 kDa
|
| Buffer: |
50 mM potassium phosphate, 135 mM NaCl, 5 mM MgCl2, pH: 6.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 9
|
Functional Validation of SAM Riboswitch Element A from Listeria monocytogenes.
Biochemistry (2024)
Hall I, Zablock K, Sobetski R, Weidmann CA, Keane SC
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human derived autoantibody mAb2G7 heavy chain, mAb2G7 VH dimer, 103 kDa protein
Human derived autoantibody mAb2G7 light chain, mAb2G7 VL dimer, 51 kDa protein
|
| Buffer: |
phosphate buffered saline, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Dec 8
|
Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat Struct Mol Biol (2024)
Wang H, Xie C, Deng B, Ding J, Li N, Kou Z, Jin M, He J, Wang Q, Wen H, Zhang J, Zhou Q, Chen S, Chen X, Yuan TF, Zhu S
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human derived autoantibody mAb2G7 heavy chain, mAb2G7 VH dimer, 103 kDa protein
Human derived autoantibody mAb2G7 light chain, mAb2G7 VL dimer, 51 kDa protein
|
| Buffer: |
phosphate buffered saline, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Dec 8
|
Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat Struct Mol Biol (2024)
Wang H, Xie C, Deng B, Ding J, Li N, Kou Z, Jin M, He J, Wang Q, Wen H, Zhang J, Zhou Q, Chen S, Chen X, Yuan TF, Zhu S
|
| RgGuinier |
5.0 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
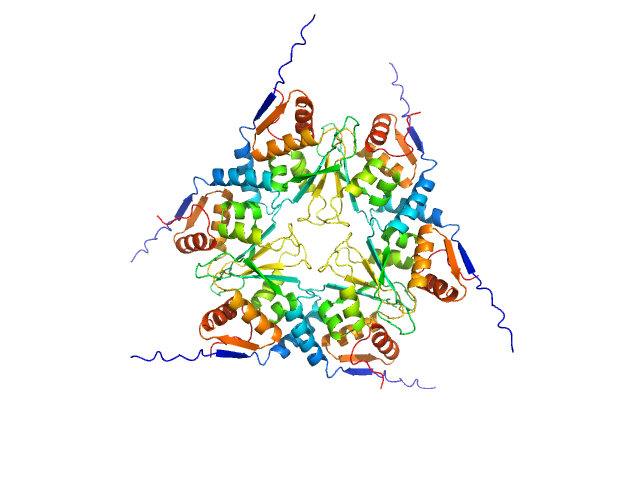
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 20
|
The Arthropoda-specific Tramtrack group BTB protein domains use previously unknown interface to form hexamers.
Elife 13 (2024)
Bonchuk AN, Balagurov KI, Baradaran R, Boyko KM, Sluchanko NN, Khrustaleva AM, Burtseva AD, Arkova OV, Khalisova KK, Popov VO, Naschberger A, Georgiev PG
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
|
|
|
|
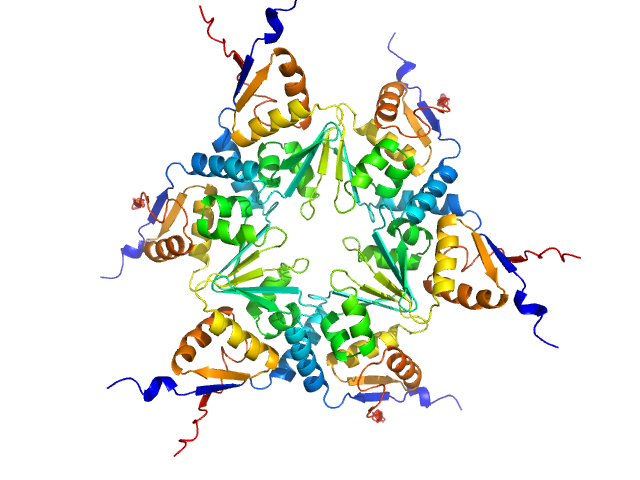
|
| Sample: |
Longitudinals lacking protein, isoform G hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 20
|
The Arthropoda-specific Tramtrack group BTB protein domains use previously unknown interface to form hexamers.
Elife 13 (2024)
Bonchuk AN, Balagurov KI, Baradaran R, Boyko KM, Sluchanko NN, Khrustaleva AM, Burtseva AD, Arkova OV, Khalisova KK, Popov VO, Naschberger A, Georgiev PG
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
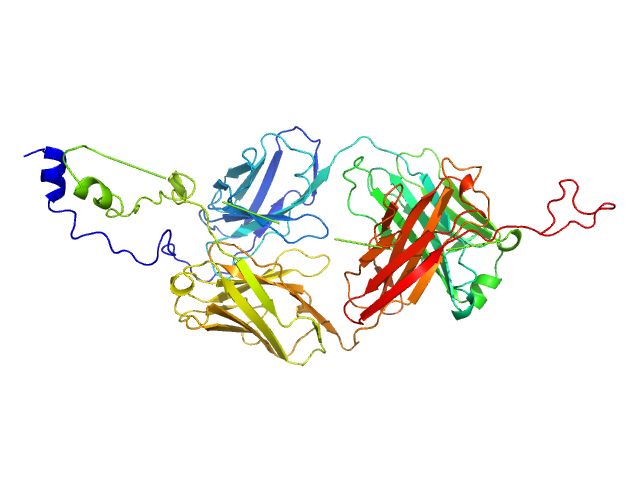
|
| Sample: |
Pro-Nivolumab, Lu02 monomer, 54 kDa protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2022 Sep 8
|
Integrating molecular dynamics simulation with small- and wide-angle X-ray scattering to unravel the flexibility, antigen-blocking, and protease-restoring functions in a hindrance-based pro-antibody.
Protein Sci 33(9):e5124 (2024)
Liao JM, Hong ST, Wang YT, Cheng YA, Ho KW, Toh SI, Shih O, Jeng US, Lyu PC, Hu IC, Huang MY, Chang CY, Cheng TL
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
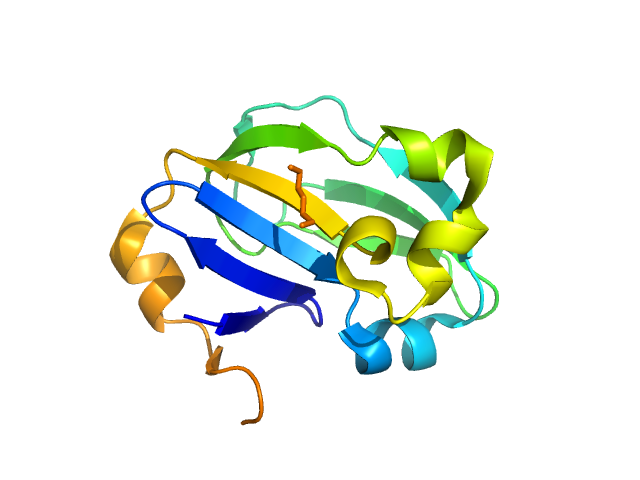
|
| Sample: |
Glycine cleavage system H-like protein monomer, 13 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
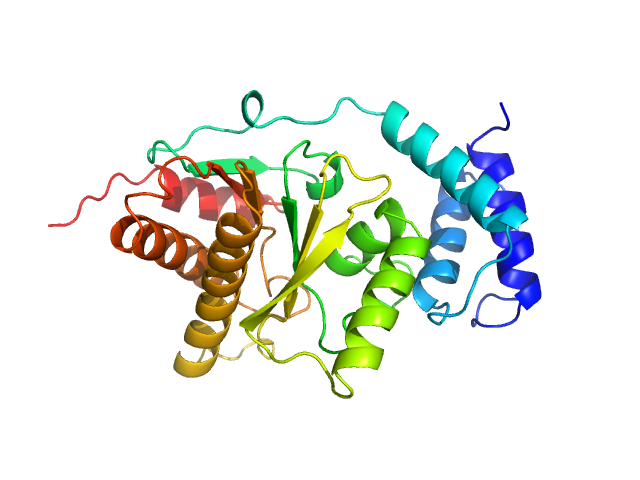
|
| Sample: |
Protein-ADP-ribose hydrolase (D13G, Y23S, T61A, I114S, R177H, I246T) monomer, 30 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
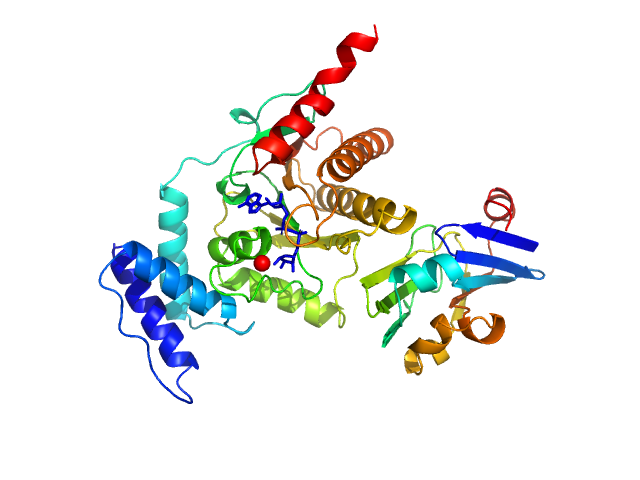
|
| Sample: |
Glycine cleavage system H-like protein monomer, 13 kDa Streptococcus pyogenes serotype … protein
Protein-ADP-ribose hydrolase (D13G, Y23S, T61A, I114S, R177H, I246T) monomer, 30 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
53 |
nm3 |
|
|