|
|
|
|
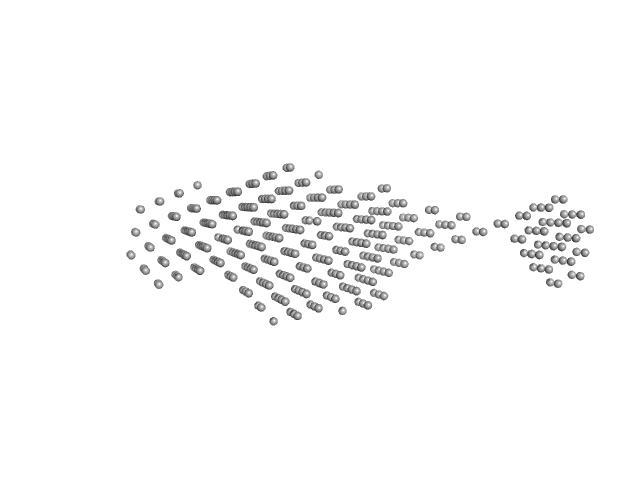
|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, 3% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Apr 24
|
LOV-activated diguanylate cyclase
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Na/Ca-exchange protein, isoform D monomer, 31 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, 1% v/v glycerol, 0.03% w/v NaN3, 1 mM β-mercaptoethanol, 2 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Mar 8
|
SAXS characterisation of CALX1.2 CBD12 construct
Roberto Kopke Salinas
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Na/Ca-exchange protein, isoform D monomer, 31 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, 1% v/v glycerol, 0.03% w/v NaN3, 1 mM β-mercaptoethanol, 50 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Mar 8
|
SAXS characterisation of CALX1.2 CBD12 construct
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
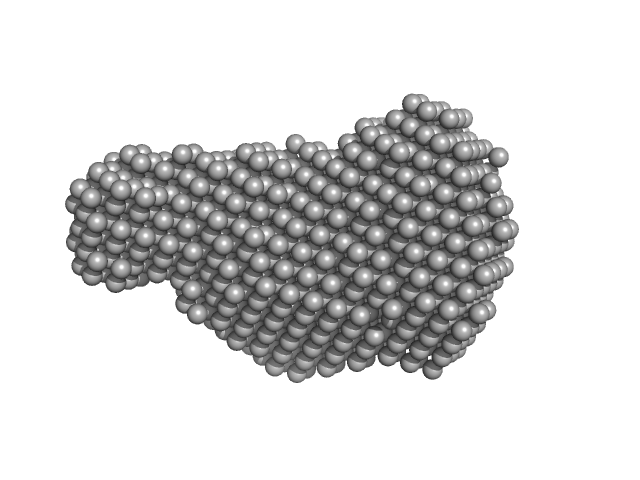
|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
MRNA-capping enzyme catalytic subunit monomer, 35 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
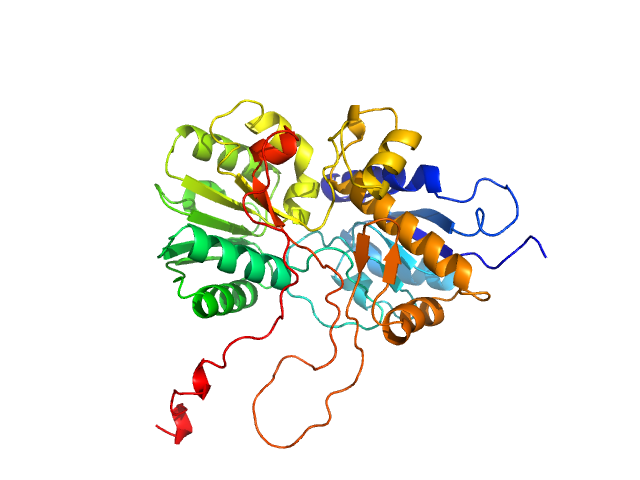
|
| Sample: |
Adenylate cyclase monomer, 43 kDa Trypanosoma brucei protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Biophysical analysis of the membrane-proximal Venus Flytrap domain of ESAG4 receptor-like adenylate cyclase from Trypanosoma brucei.
Mol Biochem Parasitol 260:111653 (2024)
Alves DO, Geens R, da Silva Arruda HR, Jennen L, Corthaut S, Wuyts E, de Andrade GC, Prosdocimi F, Cordeiro Y, Pires JR, Vieira LR, de Oliveira GAP, Sterckx YG, Salmon D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 196 kDa Bacteroides thetaiotaomicron (strain … protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Oct 22
|
BioSAXS Australian Synchrotron Standard Protein
Annmaree Warrender
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
|
|
|
|
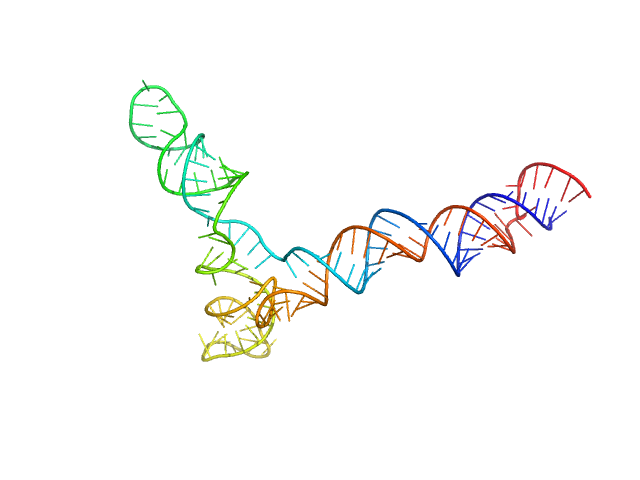
|
| Sample: |
Full stem-loop 5 of SARS-CoV-2 5'genomic end monomer, 48 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.8 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sub-element stem-loop 5a from SARS-CoV-2 5'genomic end monomer, 11 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
|
|
|
|
|
|
|
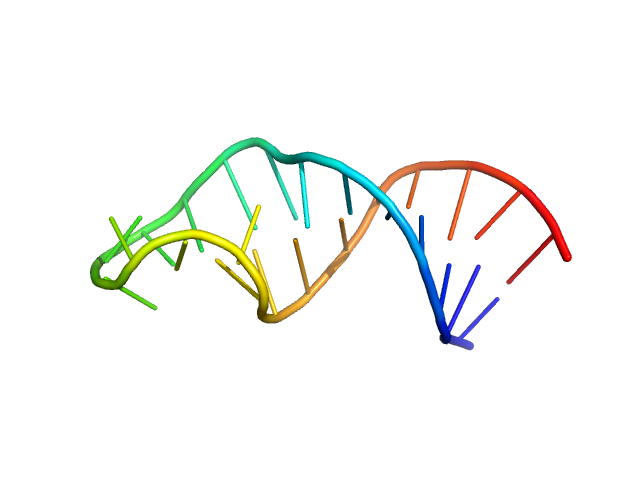
| Sample: |
Sub-element stem-loop 5b from SARS-CoV-2 5'genomic end monomer, 8 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Feb 3
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
|
|