|
|
|
|
|
| Sample: |
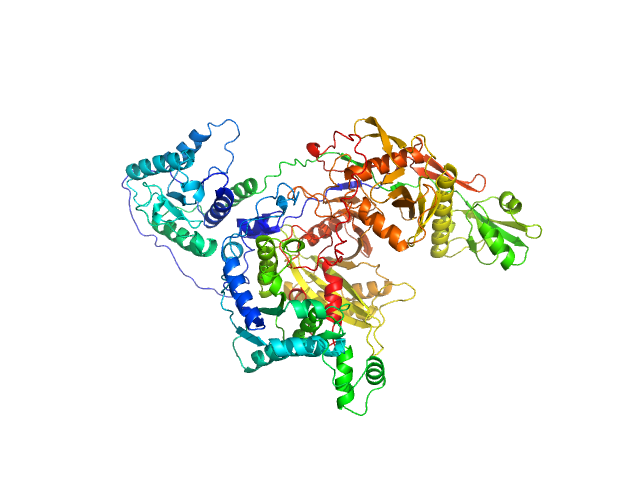
HbP1 (K526A) trimer, 56 kDa Legionella pneumophila protein
|
| Buffer: |
20 mM Tris–HCl pH 8.0, 200 mM NaCl, 5 mM EDTA, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
The Legionella collagen-like protein employs a distinct binding mechanism for the recognition of host glycosaminoglycans.
Nat Commun 15(1):4912 (2024)
Rehman S, Antonovic AK, McIntire IE, Zheng H, Cleaver L, Baczynska M, Adams CO, Portlock T, Richardson K, Shaw R, Oregioni A, Mastroianni G, Whittaker SB, Kelly G, Lorenz CD, Fornili A, Cianciotto NP, Garnett JA
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
TIR domain-containing protein monomer, 57 kDa Rhodopseudomonas palustris (strain … protein
Uncharacterized protein (PIWI) monomer, 56 kDa Rhodopseudomonas palustris (strain … protein
|
| Buffer: |
20 mM Tris-HCl, pH8.0, 200 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 27
|
Complex of Rhodopseudomonas palustris pAgo with TIR-like effector protein
Elena Manakova
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
5_SL1 monomer, 9 kDa Severe acute respiratory … RNA
|
| Buffer: |
50 mM BisTris, 25 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 1
|
The 5'-terminal stem-loop RNA element of SARS-CoV-2 features highly dynamic structural elements that are sensitive to differences in cellular pH.
Nucleic Acids Res (2024)
Toews S, Wacker A, Faison EM, Duchardt-Ferner E, Richter C, Mathieu D, Bottaro S, Zhang Q, Schwalbe H
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
1 mol% ethylammonium nitrate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 27
|
Scattering approaches to unravel protein solution behaviors in ionic liquids and deep eutectic solvents: From basic principles to recent developments
Advances in Colloid and Interface Science :103242 (2024)
Han Q, Veríssimo N, Bryant S, Martin A, Huang Y, Pereira J, Santos-Ebinuma V, Zhai J, Bryant G, Drummond C, Greaves T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
1 mol% ethylammonium nitrate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 27
|
Scattering approaches to unravel protein solution behaviors in ionic liquids and deep eutectic solvents: From basic principles to recent developments
Advances in Colloid and Interface Science :103242 (2024)
Han Q, Veríssimo N, Bryant S, Martin A, Huang Y, Pereira J, Santos-Ebinuma V, Zhai J, Bryant G, Drummond C, Greaves T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
1 mol% ethylammonium nitrate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 27
|
Scattering approaches to unravel protein solution behaviors in ionic liquids and deep eutectic solvents: From basic principles to recent developments
Advances in Colloid and Interface Science :103242 (2024)
Han Q, Veríssimo N, Bryant S, Martin A, Huang Y, Pereira J, Santos-Ebinuma V, Zhai J, Bryant G, Drummond C, Greaves T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA polymerase monomer, 117 kDa Vaccinia virus (strain … protein
DNA polymerase processivity factor component OPG148 monomer, 49 kDa Vaccinia virus (strain … protein
Uracil-DNA glycosylase monomer, 27 kDa Vaccinia virus (strain … protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl and 1 mM TCEP, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 25
|
Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
PLoS Pathog 20(5):e1011652 (2024)
Burmeister WP, Boutin L, Balestra AC, Gröger H, Ballandras-Colas A, Hutin S, Kraft C, Grimm C, Böttcher B, Fischer U, Tarbouriech N, Iseni F
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
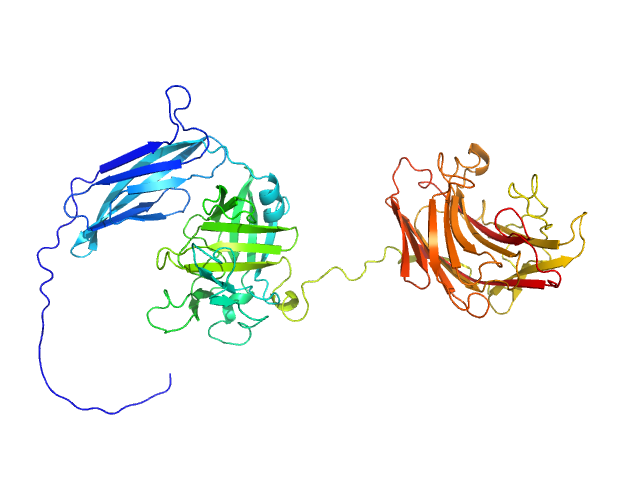
Glycosyl hydrolase, family 16 monomer, 58 kDa Christiangramia forsetii (strain … protein
|
| Buffer: |
10 mM MOPS, 100 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ Microbiol 26(5):e16624 (2024)
Zühlke MK, Ficko-Blean E, Bartosik D, Terrapon N, Jeudy A, Jam M, Wang F, Welsch N, Dürwald A, Martin LT, Larocque R, Jouanneau D, Eisenack T, Thomas F, Trautwein-Schult A, Teeling H, Becher D, Schweder T, Czjzek M
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
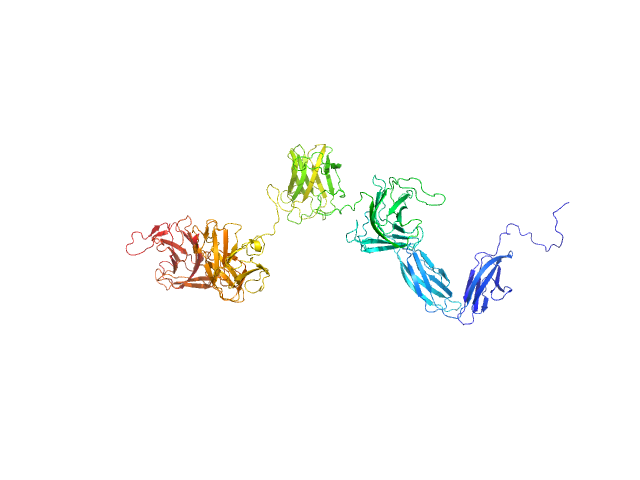
PKD domain-containing protein monomer, 96 kDa Christiangramia forsetii (strain … protein
|
| Buffer: |
10 mM MOPS, 100 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ Microbiol 26(5):e16624 (2024)
Zühlke MK, Ficko-Blean E, Bartosik D, Terrapon N, Jeudy A, Jam M, Wang F, Welsch N, Dürwald A, Martin LT, Larocque R, Jouanneau D, Eisenack T, Thomas F, Trautwein-Schult A, Teeling H, Becher D, Schweder T, Czjzek M
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Feb 15
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
253 |
nm3 |
|
|