|
|
|
|
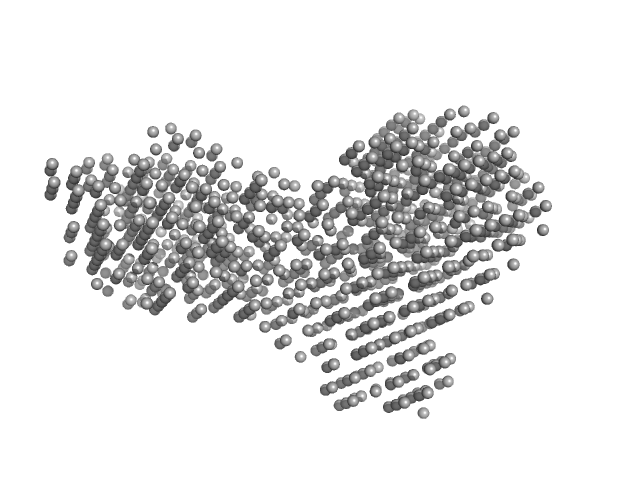
|
| Sample: |
Beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jun 4
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
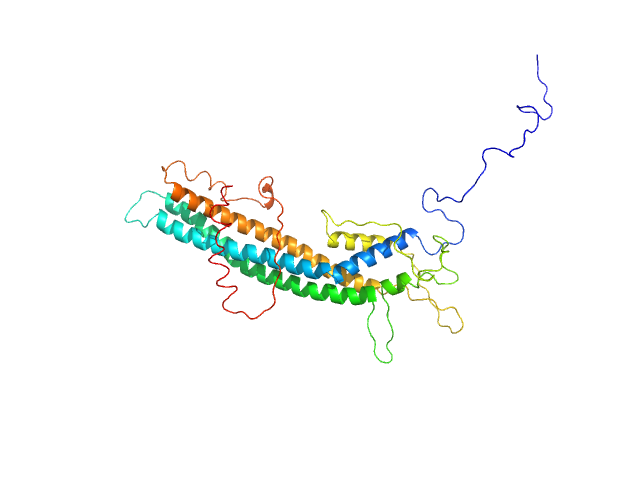
|
| Sample: |
T. brucei spp.-specific protein monomer, 38 kDa Trypanosoma brucei gambiense … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
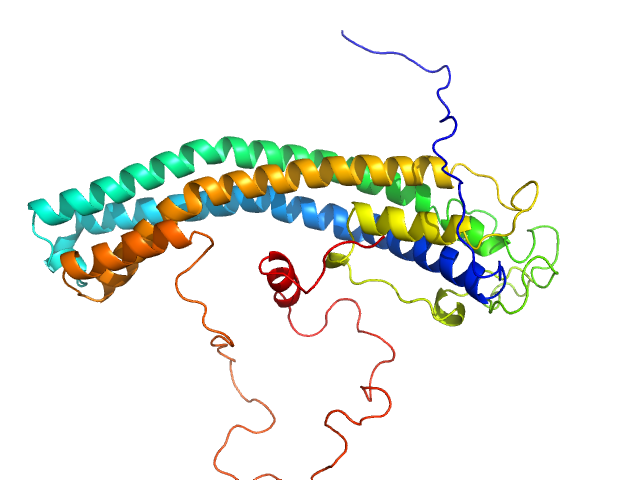
|
| Sample: |
64 kDa invariant surface glycoprotein, putative monomer, 40 kDa Trypanosoma brucei gambiense … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
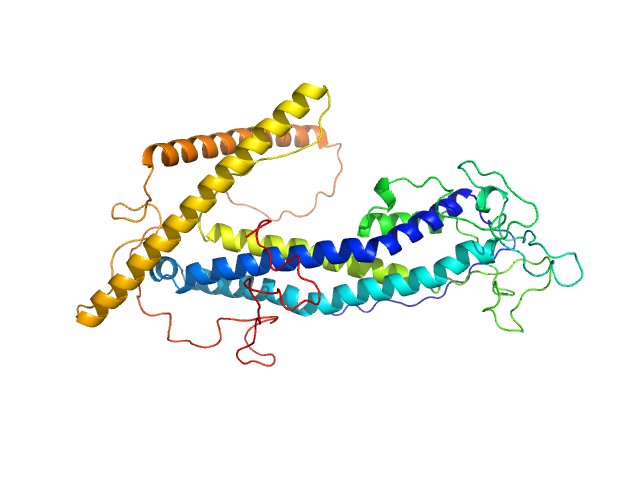
|
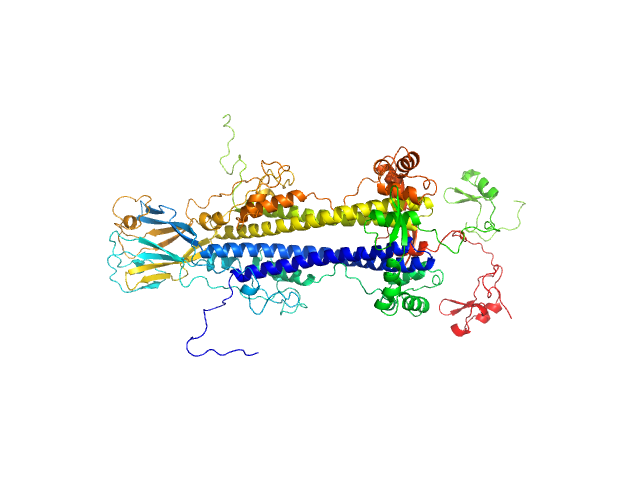
| Sample: |
ISG75 monomer, 50 kDa Trypanosoma brucei gambiense protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 25
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
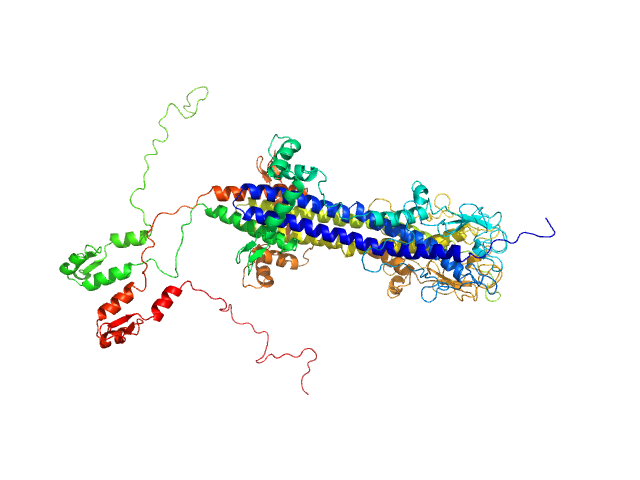
| Sample: |
Variable surface glycoprotein LiTat 1.5 dimer, 98 kDa Trypanosoma brucei gambiense protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Variant surface glycoprotein 3.1 dimer, 103 kDa Trypanosoma brucei gambiense protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
|
|
|
|
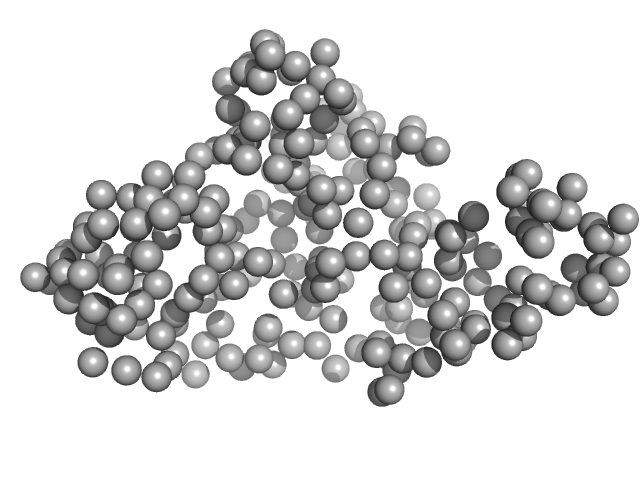
|
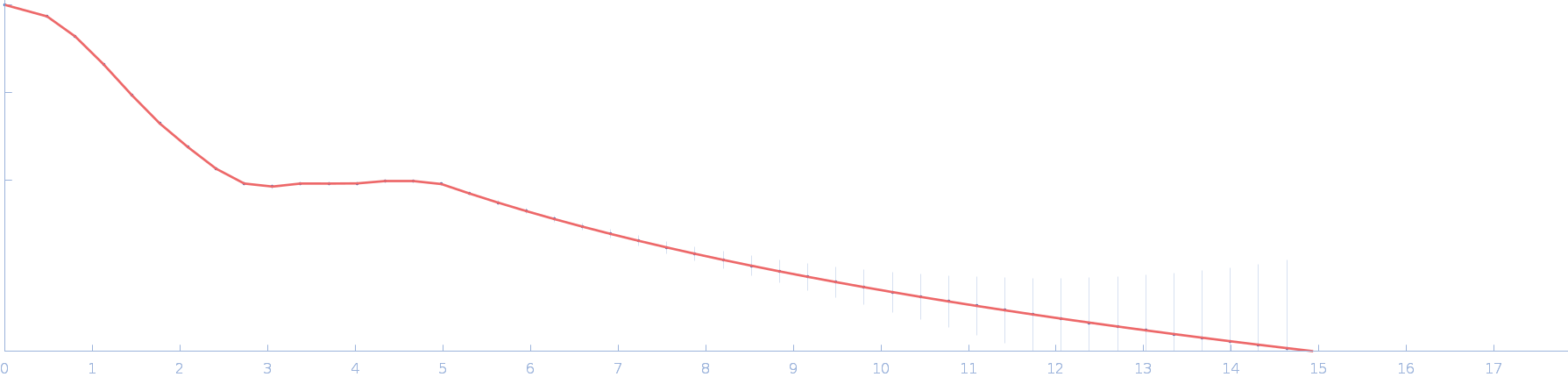
| Sample: |
RTX toxin transporter monomer, 28 kDa Kingella kingae ATCC … protein
|
| Buffer: |
100 mM HEPES, 10 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
Type 1 secretion necessitates a tight interplay between all domains of the ABC transporter
Scientific Reports 14(1) (2024)
Anlauf M, Bilsing F, Reiners J, Spitz O, Hachani E, Smits S, Schmitt L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ssr1698 protein (H21A) dimer, 22 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Dec 7
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
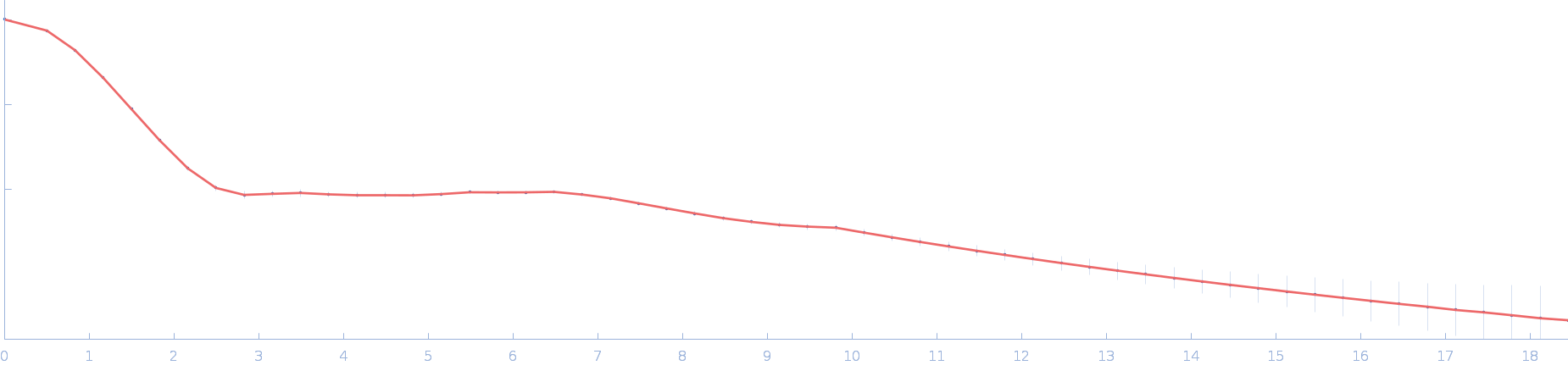
| RgGuinier |
2.0 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ssr1698 protein (H79A:R90A) dimer, 22 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Mar 30
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
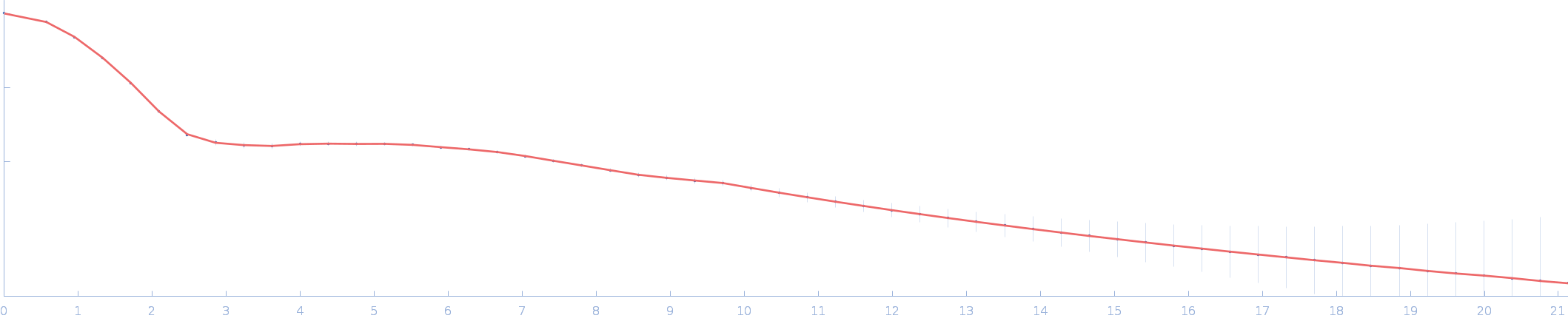
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ssr1698 protein (H79A:R90A) monomer, 11 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Mar 30
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
12 |
nm3 |
|
|