|
|
|
|
|
| Sample: |
Chitodextrinase monomer, 109 kDa Vibrio cholerae protein
|
| Buffer: |
100mM HEPES, 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2025 Apr 9
|
Multi-domain structure of a periplasmic chitodextrinase from Vibrio cholerae (VcChdx)
Kanokbhorn Khumnonkhro
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|

|
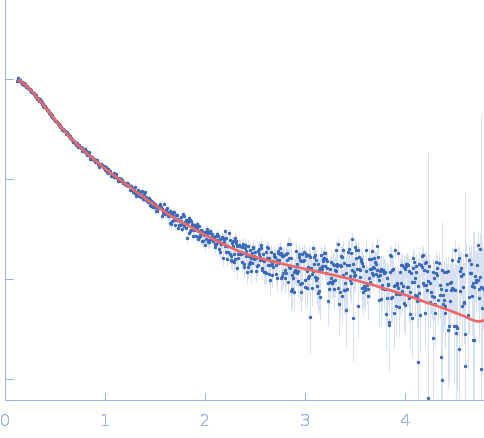
| Sample: |
S9 peptidase form Rhizobium radiobacter monomer, 74 kDa Agrobacterium radiobacter protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and functional characterization of S9 peptidase form Rhizobium radiobacter
Dr. Rahul Singh
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
137 |
nm3 |
|
|
|
|
|
|
|
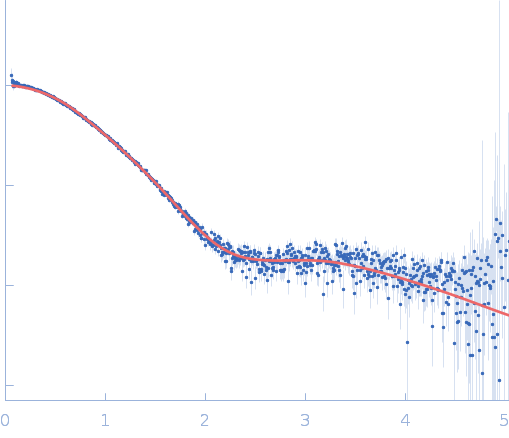
| Sample: |
S9 peptidase form Rhizobium radiobacter monomer, 74 kDa Agrobacterium radiobacter protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and functional characterization of S9 peptidase form Rhizobium radiobacter
Dr. Rahul Singh
|
| RgGuinier |
3.6 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
|
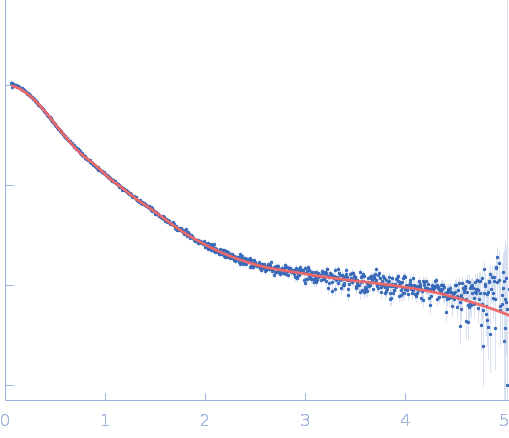
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Apr 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Jun 28
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 10 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Dec 12
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
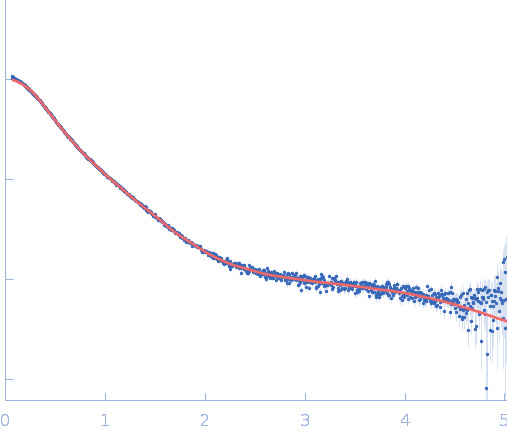
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Jun 28
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
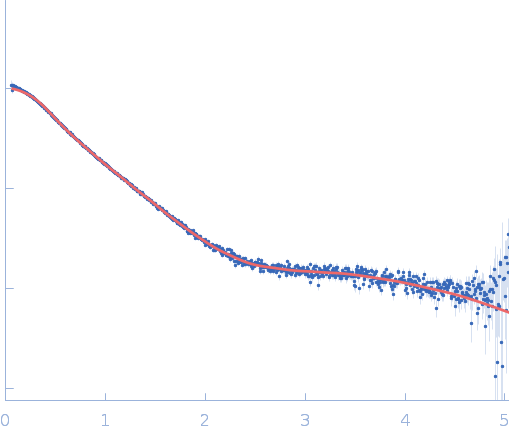
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 10 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Aug 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 1 mM SrCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Jun 28
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 2 mM SrCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Jul 22
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|