|
|
|
|
|
| Sample: |
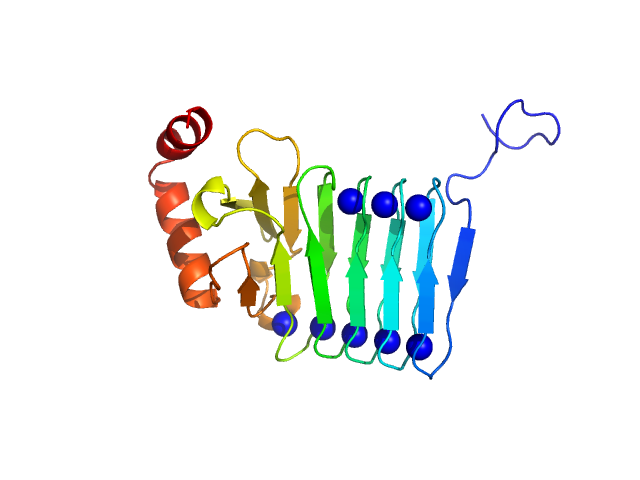
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 3 mM SrCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Jul 22
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
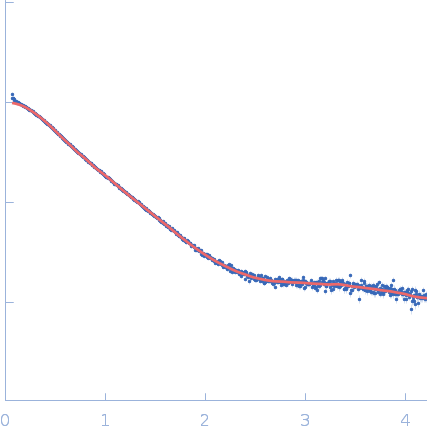
| RgGuinier |
2.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 1 mM BaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Aug 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 3 mM BaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Jul 22
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
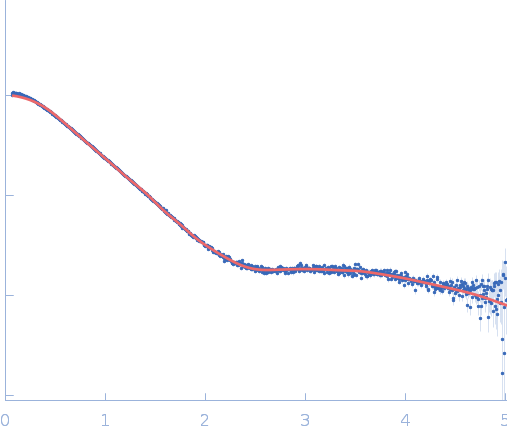
| RgGuinier |
2.5 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 10 mM BaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Aug 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
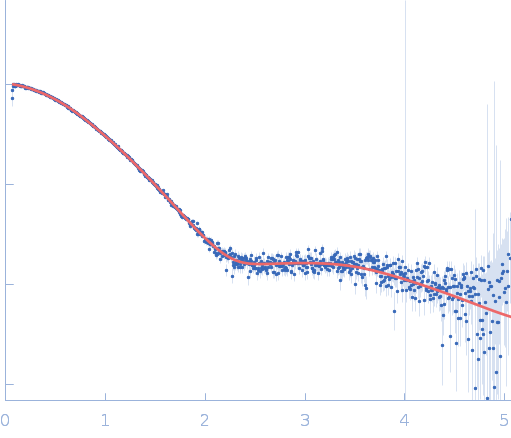
| RgGuinier |
2.7 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 10 mM SrCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Aug 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT , 3 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Dec 12
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
N-acetyl-gamma-glutamyl-phosphate reductase (ArgC) dimer, 97 kDa Paulinella chromatophora protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2023 Mar 10
|
The Paulinella
chromatophore transit peptide part2 adopts a structural fold similar to the γ-glutamyl-cyclotransferase fold
Plant Physiology (2025)
Klimenko V, Reiners J, Applegate V, Reimann K, Popowicz G, Hoeppner A, Papadopoulos A, Smits S, Nowack E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|
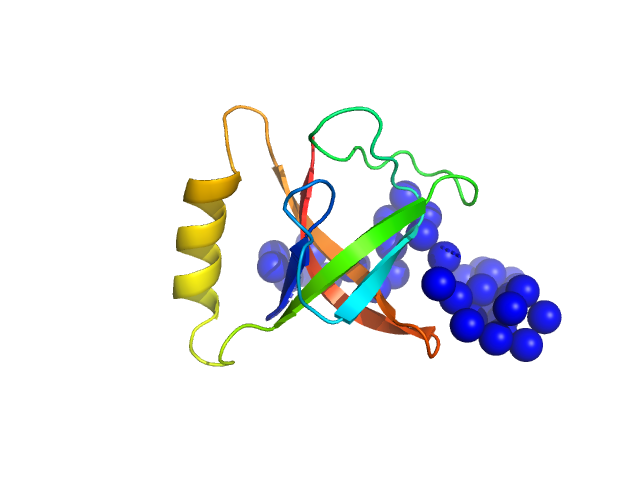
|
| Sample: |
CrTPpart2 of N-acetyl-gamma-glutamyl-phosphate reductase (ArgC) monomer, 14 kDa Paulinella chromatophora protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
The Paulinella
chromatophore transit peptide part2 adopts a structural fold similar to the γ-glutamyl-cyclotransferase fold
Plant Physiology (2025)
Klimenko V, Reiners J, Applegate V, Reimann K, Popowicz G, Hoeppner A, Papadopoulos A, Smits S, Nowack E
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNA helicase (RnaH) monomer, 63 kDa Paulinella chromatophora protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2024 Oct 28
|
The Paulinella
chromatophore transit peptide part2 adopts a structural fold similar to the γ-glutamyl-cyclotransferase fold
Plant Physiology (2025)
Klimenko V, Reiners J, Applegate V, Reimann K, Popowicz G, Hoeppner A, Papadopoulos A, Smits S, Nowack E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CrTPpart2 of RNA helicase (RnaH) monomer, 21 kDa Paulinella chromatophora protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 15
|
The Paulinella
chromatophore transit peptide part2 adopts a structural fold similar to the γ-glutamyl-cyclotransferase fold
Plant Physiology (2025)
Klimenko V, Reiners J, Applegate V, Reimann K, Popowicz G, Hoeppner A, Papadopoulos A, Smits S, Nowack E
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|