|
|
|
|
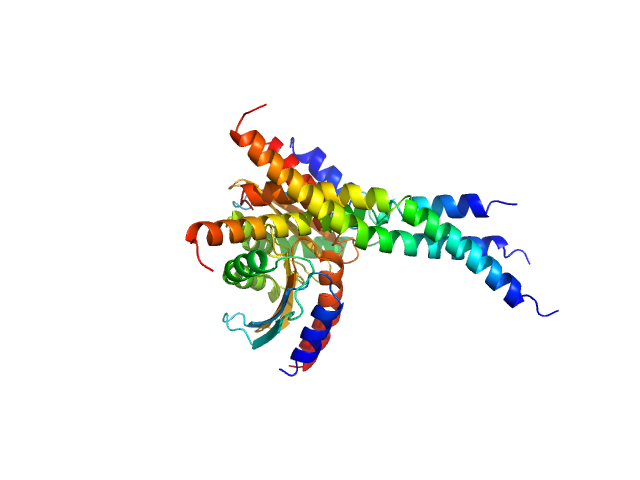
|
| Sample: |
GTPase dimer, 32 kDa Myxococcus xanthus (strain … protein
Two-component system response regulator trimer, 26 kDa Myxococcus xanthus DK … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 31
|
Structural and biophysical insights into RomR, MglB and MglC interactions involved in regulating cell polarity in Myxococcus xanthus
Journal of Biological Chemistry :110907 (2025)
Kodesia A, Kapoor S, Thakur K
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GTPase dimer, 32 kDa Myxococcus xanthus (strain … protein
Two-component system response regulator trimer, 39 kDa Myxococcus xanthus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 31
|
Structural and biophysical insights into RomR, MglB and MglC interactions involved in regulating cell polarity in Myxococcus xanthus
Journal of Biological Chemistry :110907 (2025)
Kodesia A, Kapoor S, Thakur K
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Two-component system response regulator trimer, 26 kDa Myxococcus xanthus DK … protein
Mutual gliding motility protein C dimer, 32 kDa Myxococcus xanthus (strain … protein
Gliding motility protein MglB dimer, 32 kDa Myxococcus xanthus (strain … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 31
|
Structural and biophysical insights into RomR, MglB and MglC interactions involved in regulating cell polarity in Myxococcus xanthus
Journal of Biological Chemistry :110907 (2025)
Kodesia A, Kapoor S, Thakur K
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein (Isoform P3; C297S) dimer, 56 kDa Rabies virus (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Mar 7
|
Conformational dynamics, RNA binding, and phase separation regulate the multifunctionality of rabies virus P protein.
Nat Commun 16(1):9491 (2025)
Rawlinson SM, Chakraborty S, Sethi A, David CT, Harrison AR, Bird LE, Rozario AM, Uthishtran S, Ardipradja K, Zhao T, Oksayan S, Jans DA, Ang CS, Lu ZH, Yan F, Williamson NA, Arumugam S, Sundaramoorthy V, Bell TDM, Gooley PR, Moseley GW
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein (Isoform P3; D289N, C297S) dimer, 56 kDa Rabies virus (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Mar 7
|
Conformational dynamics, RNA binding, and phase separation regulate the multifunctionality of rabies virus P protein.
Nat Commun 16(1):9491 (2025)
Rawlinson SM, Chakraborty S, Sethi A, David CT, Harrison AR, Bird LE, Rozario AM, Uthishtran S, Ardipradja K, Zhao T, Oksayan S, Jans DA, Ang CS, Lu ZH, Yan F, Williamson NA, Arumugam S, Sundaramoorthy V, Bell TDM, Gooley PR, Moseley GW
|
| RgGuinier |
3.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
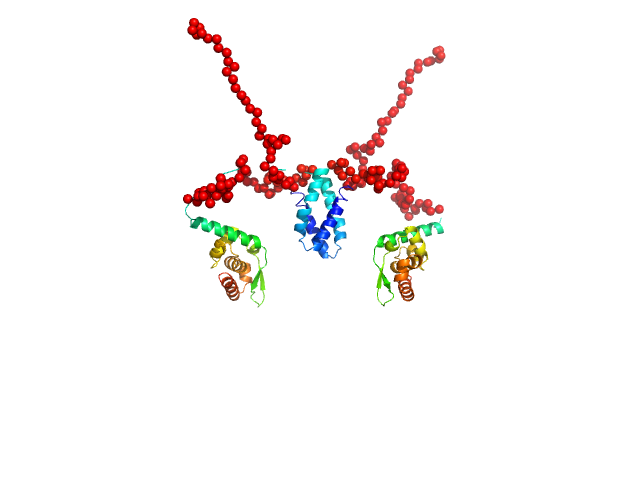
|
| Sample: |
Phosphoprotein (Isoform P3; K214A, R260A, D289N, C297S) dimer, 55 kDa Rabies virus (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Mar 7
|
Conformational dynamics, RNA binding, and phase separation regulate the multifunctionality of rabies virus P protein.
Nat Commun 16(1):9491 (2025)
Rawlinson SM, Chakraborty S, Sethi A, David CT, Harrison AR, Bird LE, Rozario AM, Uthishtran S, Ardipradja K, Zhao T, Oksayan S, Jans DA, Ang CS, Lu ZH, Yan F, Williamson NA, Arumugam S, Sundaramoorthy V, Bell TDM, Gooley PR, Moseley GW
|
| RgGuinier |
4.6 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein dimer, 67 kDa Rabies virus (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Mar 7
|
Conformational dynamics, RNA binding, and phase separation regulate the multifunctionality of rabies virus P protein.
Nat Commun 16(1):9491 (2025)
Rawlinson SM, Chakraborty S, Sethi A, David CT, Harrison AR, Bird LE, Rozario AM, Uthishtran S, Ardipradja K, Zhao T, Oksayan S, Jans DA, Ang CS, Lu ZH, Yan F, Williamson NA, Arumugam S, Sundaramoorthy V, Bell TDM, Gooley PR, Moseley GW
|
| RgGuinier |
4.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
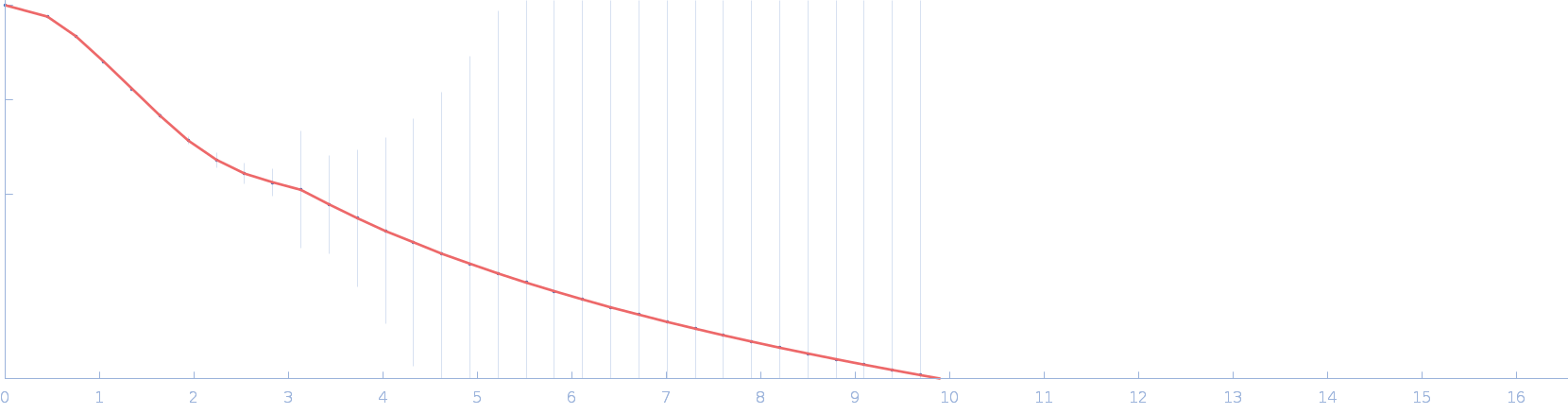
|
| Sample: |
Cellular communication network factor 2 monomer, 36 kDa protein
|
| Buffer: |
50 mM Tris pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 6
|
Structural basis for therapeutic antibody binding to CCN2 and its mechanisms on TGF-β family signaling
Haben Gabir
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
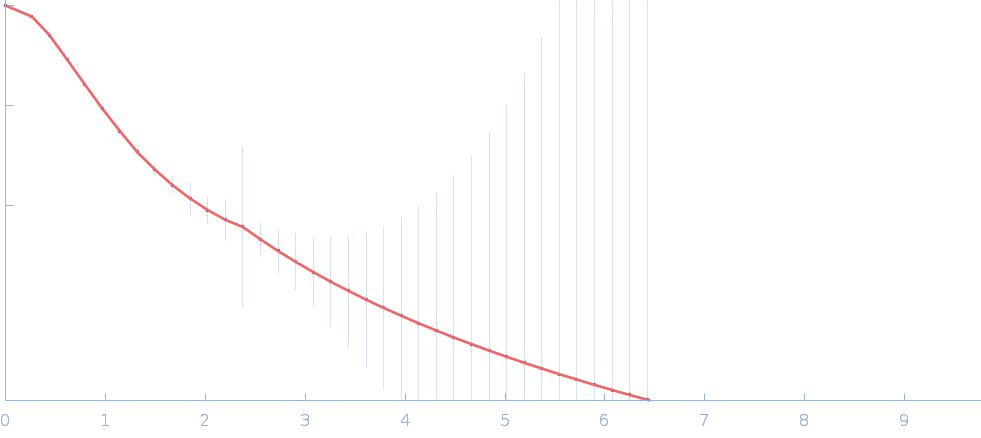
|
| Sample: |
Cellular communication network factor 2 monomer, 36 kDa protein
Anti-CCN2 37-45-MH1 F(ab) antibody monomer, 49 kDa protein
|
| Buffer: |
50 mM Tris pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 6
|
Structural basis for therapeutic antibody binding to CCN2 and its mechanisms on TGF-β family signaling
Haben Gabir
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|
|
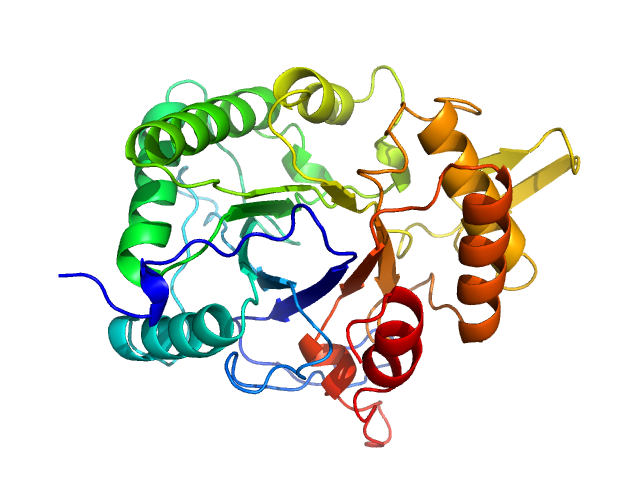
| Sample: |
Chitinase monomer, 36 kDa Listeria monocytogenes serovar … protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Aug 4
|
Structure and analysis of a virulent chitinase from Listeria monocytogenes.
Biochim Biophys Acta Proteins Proteom :141106 (2025)
Rehman S, Baczynska M, Zhang B, Lorenz CD, Garnett JA
|
| RgGuinier |
2.2 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|