|
|
|
|
|
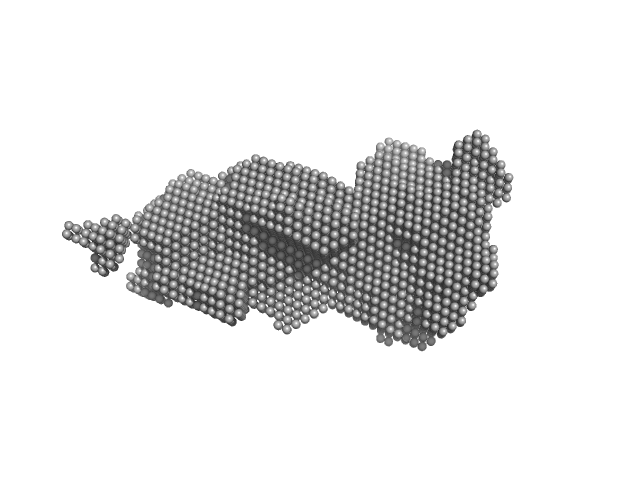
| Sample: |
Transferrin-binding protein B monomer, 32 kDa Moraxella bovis protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jun 23
|
A secreted bacterial protein protects bacteria from cationic antimicrobial peptides by entrapment in phase separated droplets
PNAS Nexus (2024)
Ostan N, Cole G, Wang F, Reichheld S, Moore G, Pan C, Yu R, Lai C, Sharpe S, Lee H, Schryvers A, Moraes T
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GGDEF domain-containing protein dimer, 132 kDa Acidimicrobiaceae bacterium protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jan 21
|
Photocobilins integrate B(12) and bilin photochemistry for enzyme control.
Nat Commun 15(1):2740 (2024)
Zhang S, Jeffreys LN, Poddar H, Yu Y, Liu C, Patel K, Johannissen LO, Zhu L, Cliff MJ, Yan C, Schirò G, Weik M, Sakuma M, Levy CW, Leys D, Heyes DJ, Scrutton NS
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GGDEF domain-containing protein dimer, 132 kDa Acidimicrobiaceae bacterium protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jan 21
|
Photocobilins integrate B(12) and bilin photochemistry for enzyme control.
Nat Commun 15(1):2740 (2024)
Zhang S, Jeffreys LN, Poddar H, Yu Y, Liu C, Patel K, Johannissen LO, Zhu L, Cliff MJ, Yan C, Schirò G, Weik M, Sakuma M, Levy CW, Leys D, Heyes DJ, Scrutton NS
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-7 monomer, 30 kDa Colletotrichum orbiculare (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-7 dimer, 60 kDa Colletotrichum orbiculare (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
4.8 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-7 monomer, 27 kDa Colletotrichum orbiculare (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Feb 1
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-7 (C286A) monomer, 30 kDa Colletotrichum orbiculare (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Feb 1
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glycosyl hydrolase family 61 monomer, 30 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 9
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glycosyl hydrolase family 61 dimer, 61 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 9
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
6.2 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-4 monomer, 33 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Feb 17
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
3.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|