|
|
|
|
|
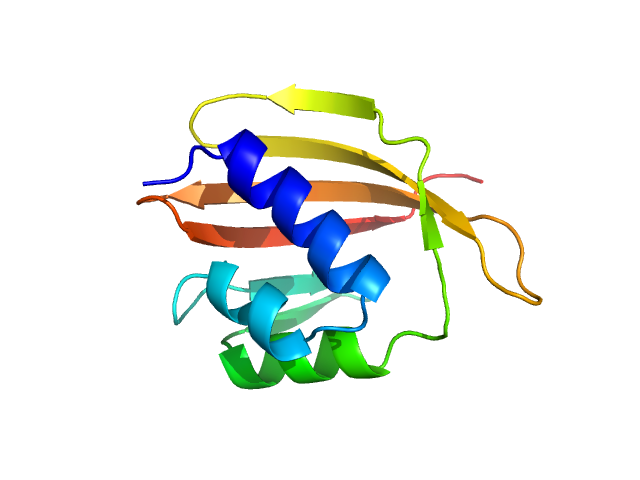
| Sample: |
Endoglucanase-4 dimer, 66 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Feb 17
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
6.4 |
nm |
| Dmax |
27.9 |
nm |
| VolumePorod |
199 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SnoaL-like domain-containing protein monomer, 12 kDa Sphaerobacter thermophilus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 6
|
Elements of the C-terminal tail of a C-terminal domain homolog of the Orange Carotenoid Protein determining xanthophyll uptake from liposomes.
Biochim Biophys Acta Bioenerg 1865(3):149043 (2024)
Likkei K, Moldenhauer M, Tavraz NN, Egorkin NA, Slonimskiy YB, Maksimov EG, Sluchanko NN, Friedrich T
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
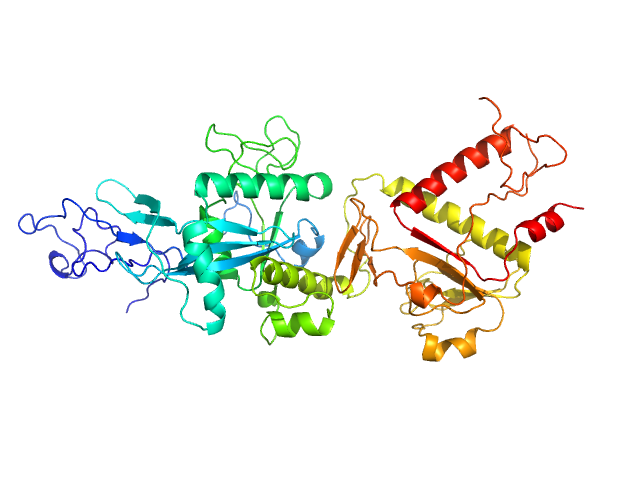
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
Replicase polyprotein 1ab (non-structural protein 16) monomer, 33 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
Replicase polyprotein 1ab (non-structural protein 16) monomer, 33 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
4.7 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|

|
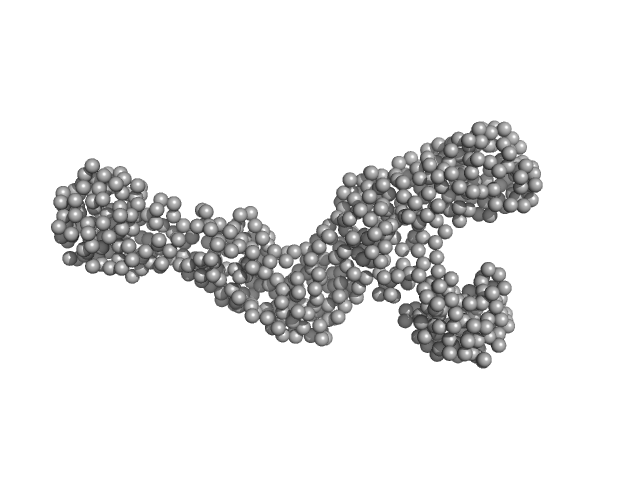
| Sample: |
Conserved protein monomer, 24 kDa Sulfolobus acidocaldarius (strain … protein
Conserved protein monomer, 44 kDa Sulfolobus acidocaldarius (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 15
|
Sequential conformational transition of ArnB, an archaeal ortholog with Sec23/Sec24 core motif
(2024)
Essen L, Korf L, Steinchen W, Watad M, Bezold F, Vogt M, Selbach L, Penner A, Tourte M, Hepp S, Albers S
|
| RgGuinier |
4.2 |
nm |
| Dmax |
19.2 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
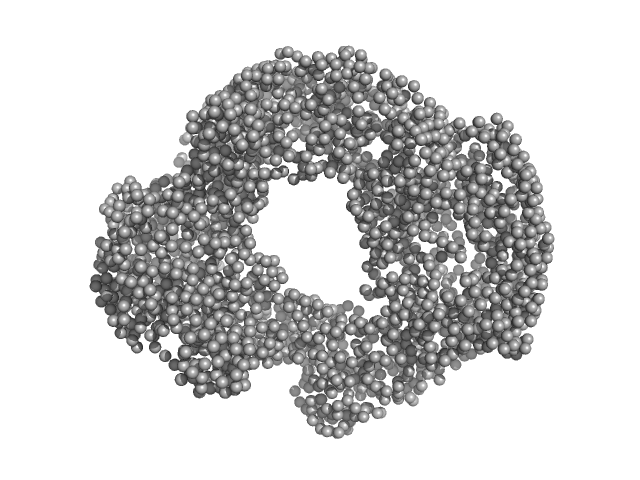
| Sample: |
Acylamino-acid-releasing enzyme (I277L, V491A) tetramer, 308 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2023 Aug 11
|
Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
FEBS Lett (2024)
Chandravanshi K, Singh R, Bhange GN, Kumar A, Yadav P, Kumar A, Makde RD
|
| RgGuinier |
5.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
424 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Invariant surface glycoprotein (N134A) monomer, 49 kDa Trypanosoma brucei protein
|
| Buffer: |
20 mM Tris-HCl, 75 mM KCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 18
|
Trypanosoma brucei Invariant Surface Glycoprotein 75 Is an Immunoglobulin Fc Receptor Inhibiting Complement Activation and Antibody-Mediated Cellular Phagocytosis.
J Immunol (2024)
Mikkelsen JH, Stødkilde K, Jensen MP, Hansen AG, Wu Q, Lorentzen J, Graversen JH, Andersen GR, Fenton RA, Etzerodt A, Thiel S, Andersen CBF
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin unc-6 monomer, 52 kDa Caenorhabditis elegans protein
Isoform a of Netrin receptor unc-5 monomer, 38 kDa Caenorhabditis elegans protein
Heparin, porcine intestinal mucosa monomer, 15 kDa Sus scrofa domesticus
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, 100 mM MgSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 18
|
Structural insights into the formation of repulsive netrin guidance complexes.
Sci Adv 10(7):eadj8083 (2024)
Priest JM, Nichols EL, Smock RG, Hopkins JB, Mendoza JL, Meijers R, Shen K, Özkan E
|
| RgGuinier |
10.4 |
nm |
| Dmax |
37.3 |
nm |
| VolumePorod |
2156 |
nm3 |
|
|