|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
200mM NaCl, 1.0mM MgCl2, 20mM Na-MES, 50µM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Jun 7
|
Wide-Angle X-ray Scattering (WAXS) Analysis of Small Nucleic Acids
Yen-Lin Chen
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
25 base-paired DNA Duplex monomer, 15 kDa DNA
|
| Buffer: |
100 mM NaCl, 10 mM sodium 3-(N-morpholino)propanesulfonic acid (Na-MOPS) and 20 uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Jun 7
|
Wide-Angle X-ray Scattering (WAXS) Analysis of Small Nucleic Acids
Yen-Lin Chen
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
200mM NaCl, 1.0mM MgCl2, 20mM Na-MES, 50µM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Jun 7
|
Wide-Angle X-ray Scattering (WAXS) Analysis of Small Nucleic Acids
Yen-Lin Chen
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Triplex monomer, 17 kDa RNA
|
| Buffer: |
200mM NaCl, 1.0mM MgCl2, 20mM Na-MES, 50µM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
Wide-Angle X-ray Scattering (WAXS) Analysis of Small Nucleic Acids
Yen-Lin Chen
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
30nt Single-stranded RNA monomer, 10 kDa RNA
|
| Buffer: |
100 mM NaCl, 10 mM sodium 3-(N-morpholino)propanesulfonic acid (Na-MOPS) and 20 uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
Wide-Angle X-ray Scattering (WAXS) Analysis of Small Nucleic Acids
Yen-Lin Chen
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Beta-galactosidase-like enzyme dimer, 129 kDa Hamamotoa singularis protein
|
| Buffer: |
5 mM sodium phosphate, pH: 5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Oak Ridge National Laboratory on 2018 Jun 25
|
Structural analysis and functional evaluation of the disordered ß–hexosyltransferase region from Hamamotoa (Sporobolomyces) singularis
Frontiers in Bioengineering and Biotechnology 11 (2023)
Dagher S, Vaishnav A, Stanley C, Meilleur F, Edwards B, Bruno-Bárcena J
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-galactosidase-like enzyme dimer, 129 kDa Hamamotoa singularis protein
|
| Buffer: |
5 mM sodium phosphate, pH: 5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Oak Ridge National Laboratory on 2018 Jun 25
|
Structural analysis and functional evaluation of the disordered ß–hexosyltransferase region from Hamamotoa (Sporobolomyces) singularis
Frontiers in Bioengineering and Biotechnology 11 (2023)
Dagher S, Vaishnav A, Stanley C, Meilleur F, Edwards B, Bruno-Bárcena J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Circumsporozoite protein monomer, 11 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
50 mM MES, 500 mM NaCl, 1 mM TCEP, pH: 6.6 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Biophysical characterization of the Plasmodium falciparum
circumsporozoite protein's N‐terminal domain
Protein Science (2023)
Geens R, Stanisich J, Beyens O, D'Hondt S, Thiberge J, Ryckebosch A, Groot A, Magez S, Vertommen D, Amino R, Winter H, Volkov A, Tompa P, Sterckx Y
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
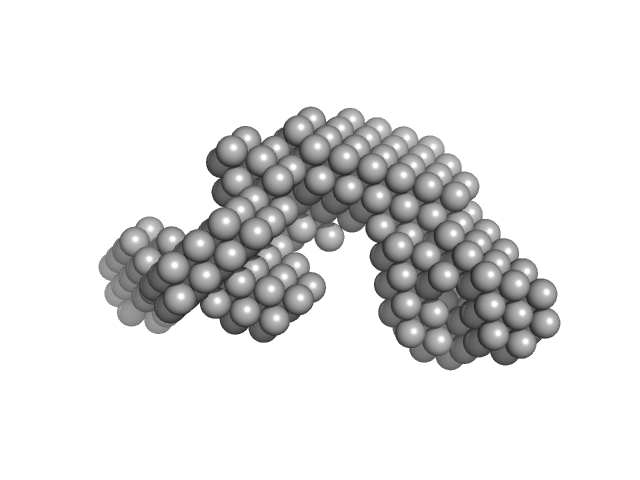
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
296 |
nm3 |
|
|
|
|
|
|
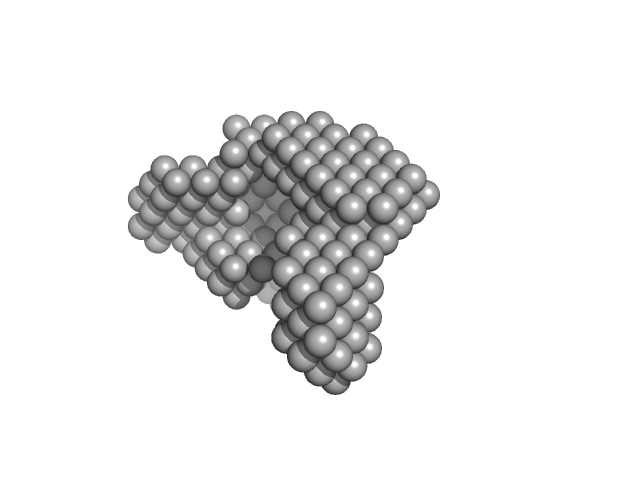
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 336-418 monomer, 9 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
279 |
nm3 |
|
|