|
|
|
|
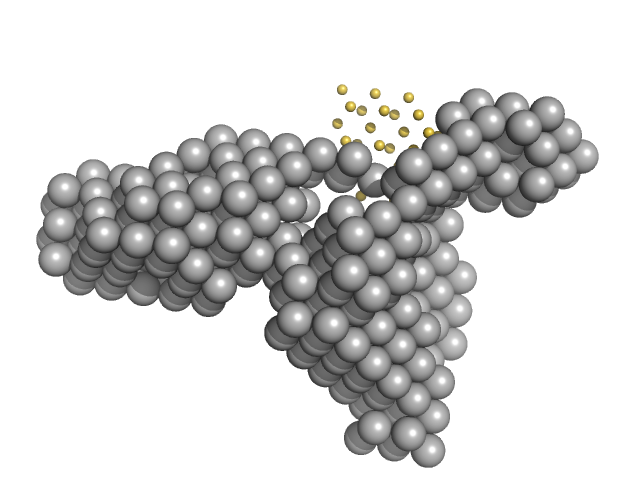
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 225-418 monomer, 21 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
|
|
|
|
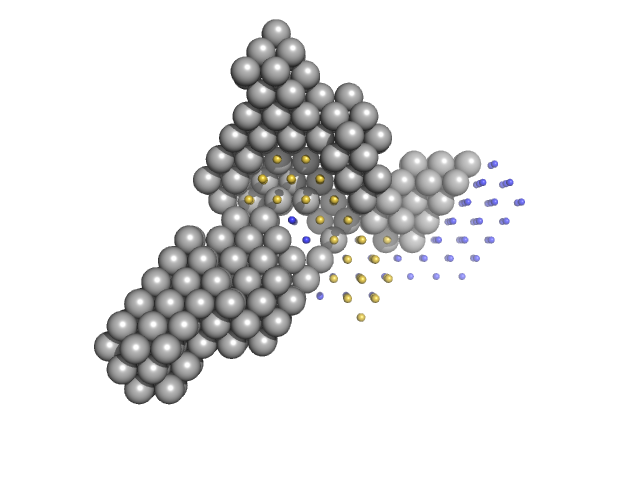
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 225-418 monomer, 21 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 776-898 monomer, 14 kDa Chaetomium thermophilum protein
SMC hinge domain-containing protein, 263-466 monomer, 22 kDa Chaetomium thermophilum protein
SMC hinge domain-containing protein, 1367-1542 monomer, 20 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
355 |
nm3 |
|
|
|
|
|
|
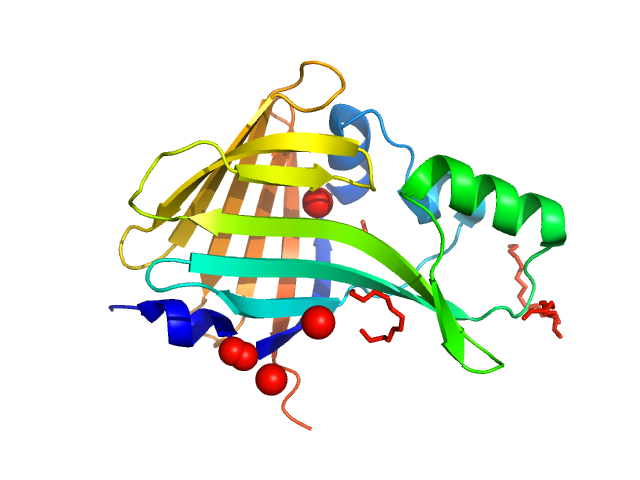
|
| Sample: |
Oplophorus-luciferin 2-monooxygenase catalytic subunit monomer, 20 kDa Oplophorus gracilirostris protein
|
| Buffer: |
10 mM Tris-HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2021 May 27
|
Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun 14(1):7864 (2023)
Nemergut M, Pluskal D, Horackova J, Sustrova T, Tulis J, Barta T, Baatallah R, Gagnot G, Novakova V, Majerova M, Sedlackova K, Marques SM, Toul M, Damborsky J, Prokop Z, Bednar D, Janin YL, Marek M
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
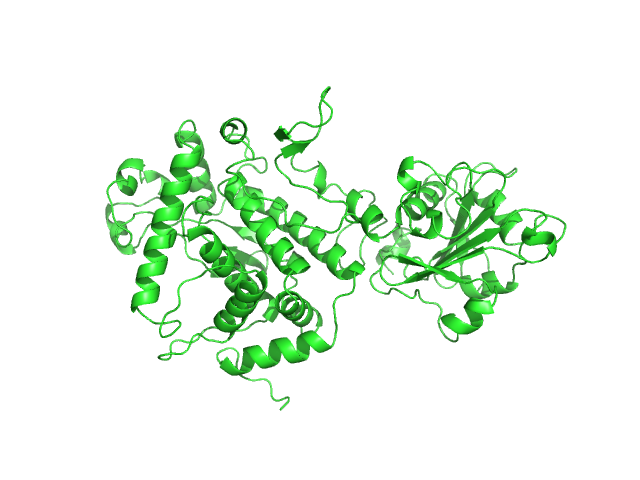
|
| Sample: |
Apoptosis inducing protein monomer, 57 kDa Photobacterium damselae subsp. … protein
|
| Buffer: |
50 mM Hepes, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Apr 13
|
Unconventional structure and mechanisms for membrane interaction and translocation of the NF-κB-targeting toxin AIP56.
Nat Commun 14(1):7431 (2023)
Lisboa J, Pereira C, Pinto RD, Rodrigues IS, Pereira LMG, Pinheiro B, Oliveira P, Pereira PJB, Azevedo JE, Durand D, Benz R, do Vale A, Dos Santos NMS
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ribosome maturation factor RimP monomer, 18 kDa Staphylococcus aureus (strain … protein
30S ribosomal protein S12 monomer, 15 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
50 mM sodium phosphate, 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at Xeuss 3.0 SAXS/WAXS System, JINR on 2023 Feb 16
|
Structural aspects of RimP binding on small ribosomal subunit from Staphylococcus aureus.
Structure (2023)
Garaeva N, Fatkhullin B, Murzakhanov F, Gafurov M, Golubev A, Bikmullin A, Glazyrin M, Kieffer B, Jenner L, Klochkov V, Aganov A, Rogachev A, Ivankov O, Validov S, Yusupov M, Usachev K
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
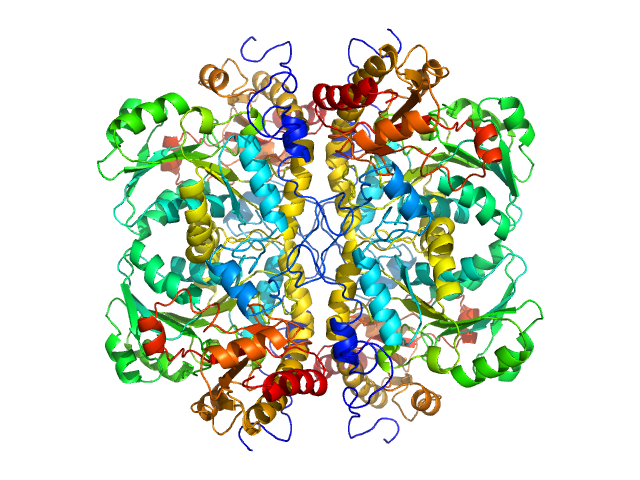
| Sample: |
L-methionine gamma-lyase tetramer, 181 kDa Clostridium tetani protein
|
| Buffer: |
PBS-D2O: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (D2O buffer), pH: 7.4 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 May 19
|
Methionine gamma lyase fused with S3 domain VGF forms octamers and adheres to tumor cells via binding EGFR
Biochemical and Biophysical Research Communications :149319 (2023)
Bondarev N, Bagaeva D, Bazhenov S, Buben M, Bulushova N, Ryzhykau Y, Okhrimenko I, Zagryadskaya Y, Maslov I, Anisimova N, Sokolova D, Kuklin A, Pokrovsky V, Manukhov I
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
232 |
nm3 |
|
|
|
|
|
|
|
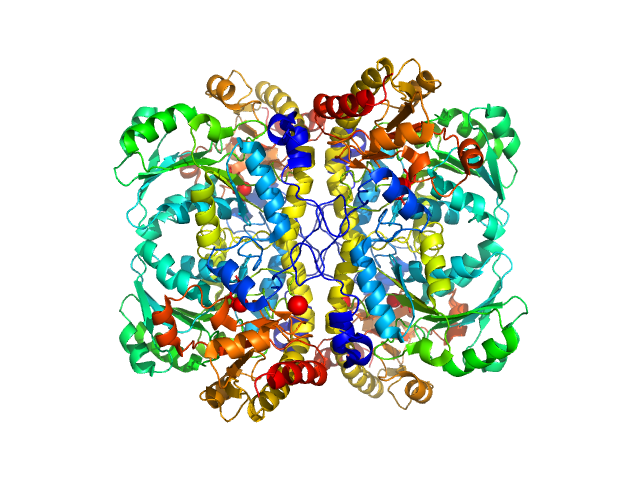
| Sample: |
L-methionine gamma-lyase (K272S) tetramer, 174 kDa Clostridium sporogenes protein
|
| Buffer: |
PBS-D2O: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (D2O buffer), pH: 7.4 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 May 19
|
Methionine gamma lyase fused with S3 domain VGF forms octamers and adheres to tumor cells via binding EGFR
Biochemical and Biophysical Research Communications :149319 (2023)
Bondarev N, Bagaeva D, Bazhenov S, Buben M, Bulushova N, Ryzhykau Y, Okhrimenko I, Zagryadskaya Y, Maslov I, Anisimova N, Sokolova D, Kuklin A, Pokrovsky V, Manukhov I
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
|
|
|
|
|
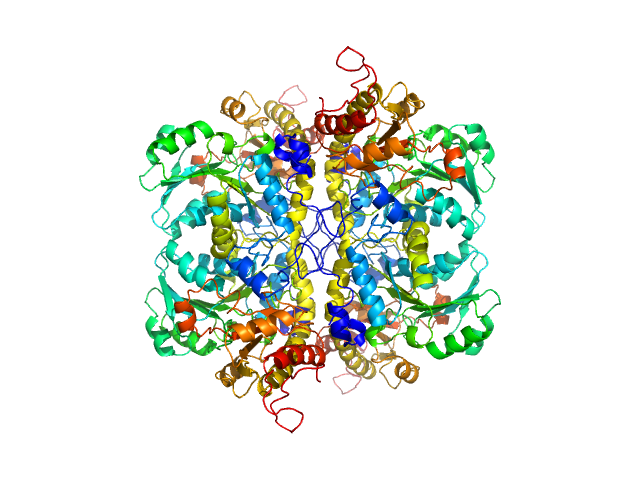
| Sample: |
L-methionine gamma-lyase from Clostridium sporogenes fused with VGF S3 domain tetramer, 183 kDa Clostridium sporogenes protein
|
| Buffer: |
PBS-D2O: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (D2O buffer), pH: 7.4 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 May 19
|
Methionine gamma lyase fused with S3 domain VGF forms octamers and adheres to tumor cells via binding EGFR
Biochemical and Biophysical Research Communications :149319 (2023)
Bondarev N, Bagaeva D, Bazhenov S, Buben M, Bulushova N, Ryzhykau Y, Okhrimenko I, Zagryadskaya Y, Maslov I, Anisimova N, Sokolova D, Kuklin A, Pokrovsky V, Manukhov I
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.3 |
nm |
| VolumePorod |
303 |
nm3 |
|
|
|
|
|
|
|
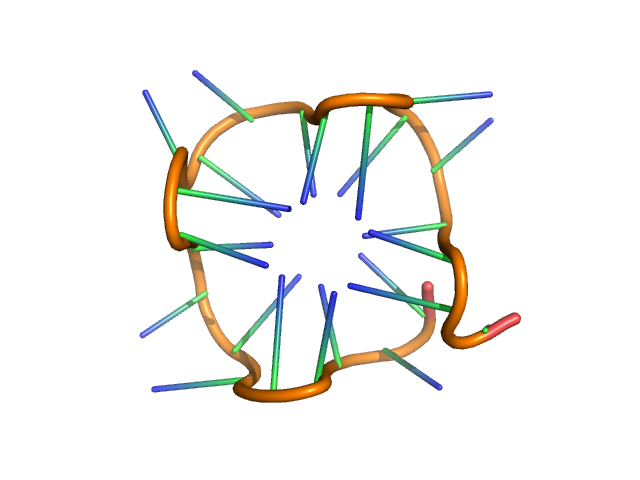
| Sample: |
PolyGU RNA - (GU)12 monomer, 8 kDa synthetic RNA RNA
|
| Buffer: |
20 mM HEPES, 150 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 10
|
Solution structure of poly(UG) RNA
Journal of Molecular Biology :168340 (2023)
Escobar C, Petersen R, Tonelli M, Fan L, Henzler-Wildman K, Butcher S
|
| RgGuinier |
1.4 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
|
|
|
|
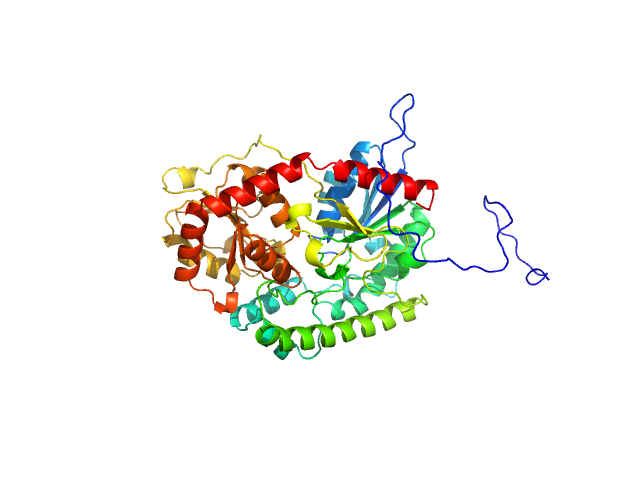
|
| Sample: |
UDP-glycosyltransferase 202A2 monomer, 53 kDa Tetranychus urticae protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 17
|
Structural and functional studies reveal the molecular basis of substrate promiscuity of a glycosyltransferase originating from a major agricultural pest
Journal of Biological Chemistry :105421 (2023)
Arriaza R, Abiskaroon B, Patel M, Daneshian L, Kluza A, Snoeck S, Watkins M, Hopkins J, Van Leeuwen T, Grbic M, Grbic V, Borowski T, Chruszcz M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
85 |
nm3 |
|
|