|
|
|
|

|
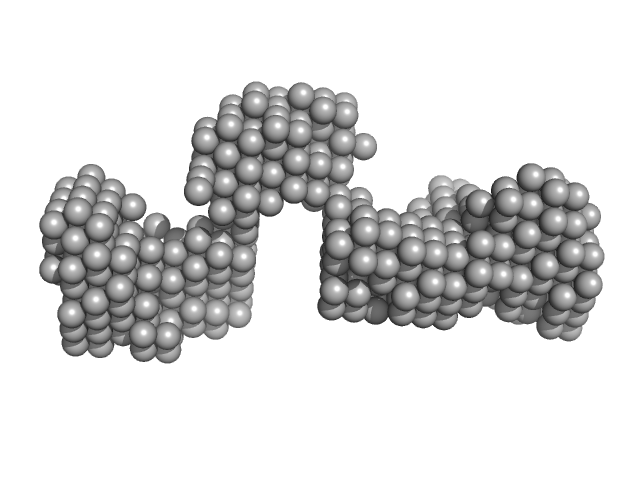
| Sample: |
Heme acquisition protein HasAp dimer, 38 kDa Pseudomonas fluorescens (strain … protein
|
| Buffer: |
50 mM CHES, 5 % glycerol, pH: 9.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2023 Feb 24
|
Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom
RSC Advances 14(13):8829-8836 (2024)
Inaba H, Shisaka Y, Ariyasu S, Sakakibara E, Ueda G, Aiba Y, Shimizu N, Sugimoto H, Shoji O
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SBV 3'UTR monomer, 31 kDa Sacbrood virus RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Apr 28
|
Biophysical characterization of honey bee virus untranslated regions
University of Lethbridge Dept. of Chemistry and Biochemistry Thesis (2024)
Jenna M Letain, Trushar R Patel
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
SBV 5'UTR monomer, 61 kDa Sacbrood virus RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 19
|
Biophysical characterization of honey bee virus untranslated regions
University of Lethbridge Dept. of Chemistry and Biochemistry Thesis (2024)
Jenna M Letain, Trushar R Patel
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
|
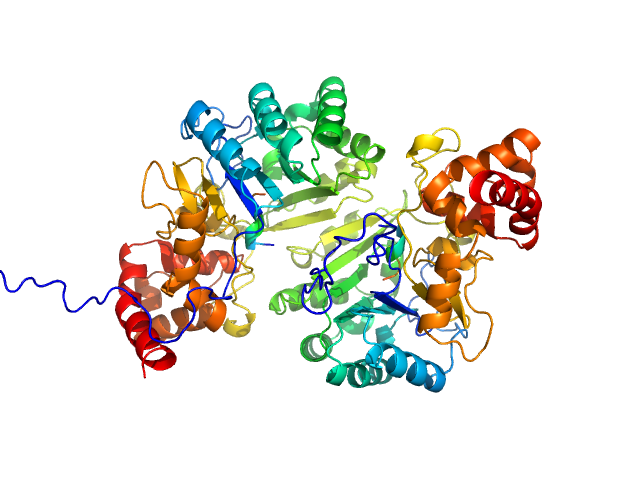
| Sample: |
Putative peptide biosynthesis protein YydG dimer, 80 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Nov 30
|
Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat Chem Biol (2023)
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
|
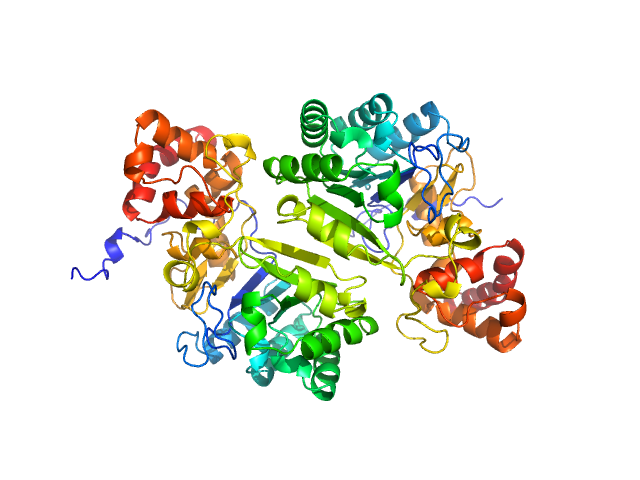
| Sample: |
Putative peptide biosynthesis protein YydG dimer, 80 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Nov 29
|
Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat Chem Biol (2023)
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HBV G4 WT monomer, 7 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 15
|
HBV G-quad
Trushar Patel
|
| RgGuinier |
1.8 |
nm |
| Dmax |
4.7 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HBV G-quad G1738A monomer, 7 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 14
|
HBV G-quad
Trushar Patel
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.3 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HBV G4 G1748A monomer, 7 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 14
|
HBV G-quad
Trushar Patel
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HBV G4 G1748A + G1748A monomer, 7 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 17
|
HBV G-quad
Trushar Patel
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
25 base-paired DNA Duplex monomer, 15 kDa DNA
|
| Buffer: |
100 mM NaCl, 10 mM sodium 3-(N-morpholino)propanesulfonic acid (Na-MOPS) and 20 uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Jun 7
|
Wide-Angle X-ray Scattering (WAXS) Analysis of Small Nucleic Acids
Yen-Lin Chen
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.4 |
nm |
|
|