|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20 mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 1mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 9
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20 mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 9
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20 mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 3 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 9
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 3.75 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 9
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20 mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 0.25 mM MgCl2 SAM, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 10
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20 mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 1 mM MgCl2 SAM, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 10
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20 mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 2 mM MgCl2 SAM, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 10
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20 mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 3 mM MgCl2 SAM, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 10
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
| Sample: |
S-adenosylmethionine riboswitch II monomer, 17 kDa RNA
|
| Buffer: |
20 mM NaMOPS, 20 mM NaCl, 20 μM EDTA, 3.75 mM MgCl2 SAM, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Dec 10
|
SAXS Measurements of the SAM2 Riboswitch
Alex Plumridge
|
|
|
|
|
|
|
|
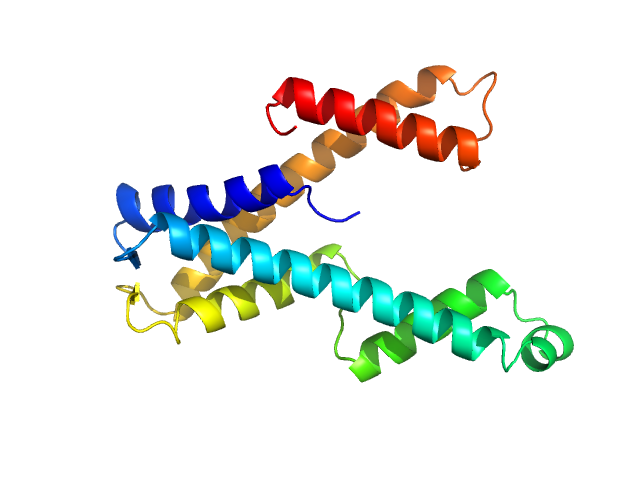
| Sample: |
DUF507 family protein monomer, 22 kDa Aquifex aeolicus (strain … protein
|
| Buffer: |
20 mM Hepes pH 7.2, 400 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 7
|
Crystal structure of domain of unknown function 507 (DUF507) reveals a new protein fold.
Sci Rep 13(1):13496 (2023)
McKay CE, Cheng J, Tanner JJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|