|
|
|
|
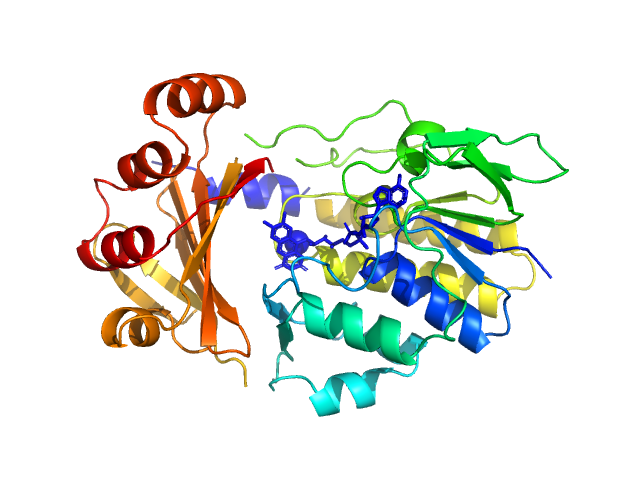
|
| Sample: |
Monooxygenase (M154I, A283T) monomer, 39 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jan 10
|
Tetracycline-modifying enzyme Sm
TetX from Stenotrophomonas maltophilia
Acta Crystallographica Section F Structural Biology Communications 79(7):180-192 (2023)
Malý M, Kolenko P, Stránský J, Švecová L, Dušková J, Koval' T, Skálová T, Trundová M, Adámková K, Černý J, Božíková P, Dohnálek J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
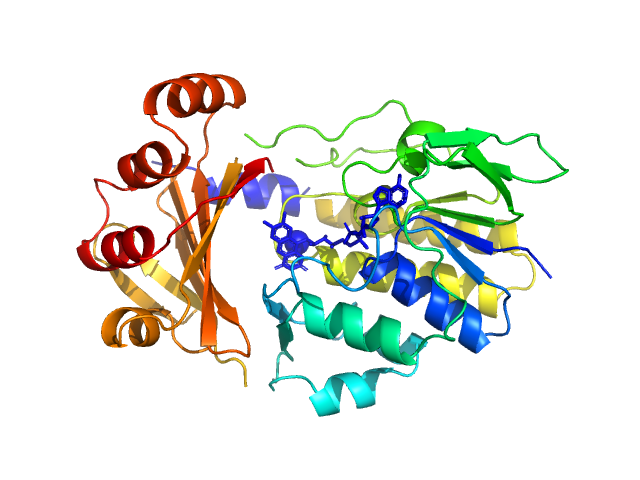
|
| Sample: |
Monooxygenase (M154I, A283T) monomer, 39 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 30 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jan 7
|
Tetracycline-modifying enzyme Sm
TetX from Stenotrophomonas maltophilia
Acta Crystallographica Section F Structural Biology Communications 79(7):180-192 (2023)
Malý M, Kolenko P, Stránský J, Švecová L, Dušková J, Koval' T, Skálová T, Trundová M, Adámková K, Černý J, Božíková P, Dohnálek J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
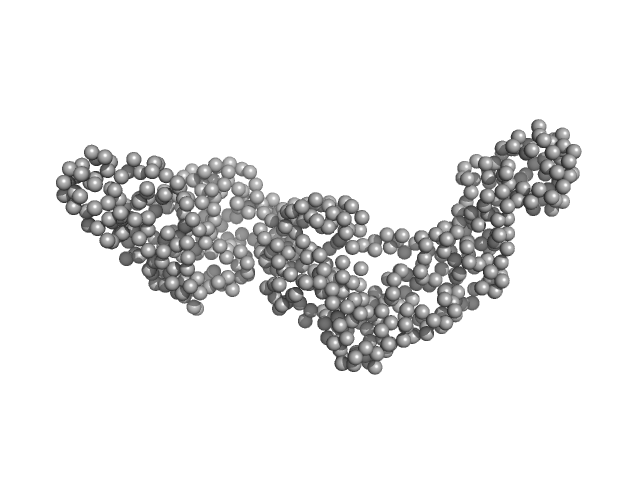
|
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2020 May 29
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
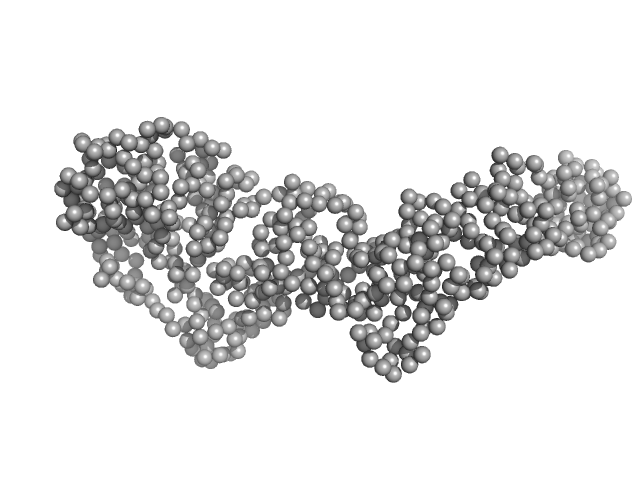
|
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
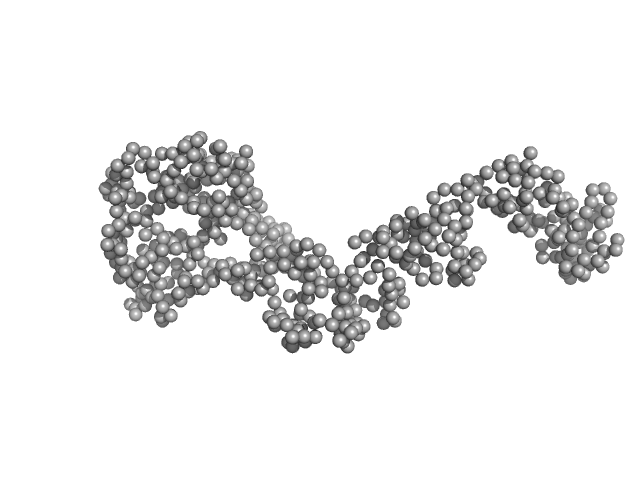
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
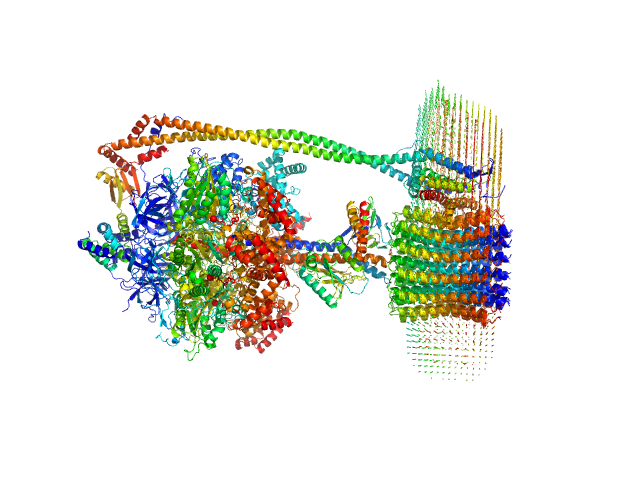
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
300 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
6.6 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
927 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRS8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
150 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
9.6 |
nm |
| Dmax |
41.5 |
nm |
| VolumePorod |
1506 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRT8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
250 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
7.4 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
949 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRU8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
300 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
39.5 |
nm |
| VolumePorod |
1127 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRV8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
350 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
8.9 |
nm |
| Dmax |
44.5 |
nm |
| VolumePorod |
1150 |
nm3 |
|
|