|
|
|
|
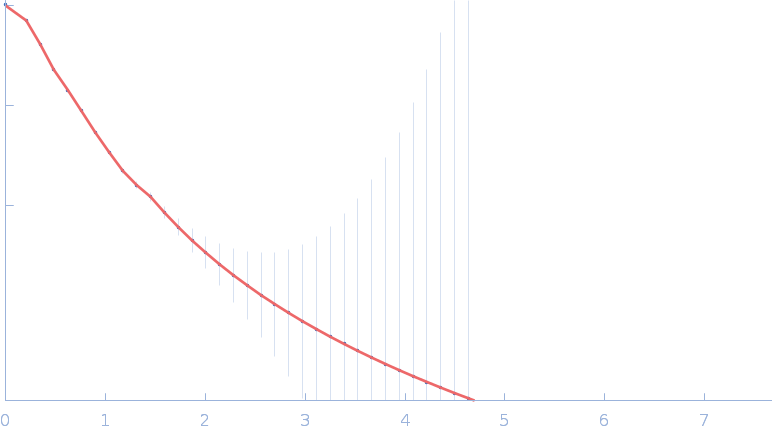
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 28 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
3.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
4.0 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 37 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 33 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
2.5 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 32 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 29 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
2.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 15 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|