|
|
|
|
|
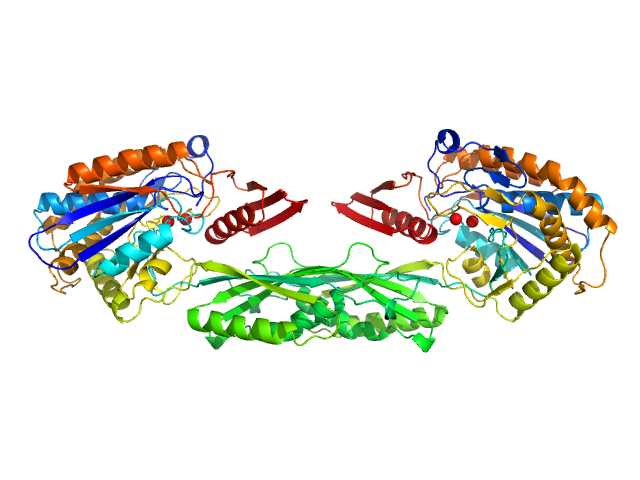
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 Disulfide Variant dimer, 96 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (HIV-1 reverse transcriptase C38V/C280S, L289K truncated to 1 - 556) monomer, 65 kDa Human immunodeficiency virus … protein
|
| Buffer: |
50 mM Bis-Tris pH 6.8, 100 mM NaCl, 1% glycerol, 0.02% w/v NaN3, pH: 6.8
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jun 20
|
Relative domain orientation of the L289K HIV
‐1 reverse transcriptase monomer
Protein Science 31(5) (2022)
Xi Z, Ilina T, Guerrero M, Fan L, Sluis‐Cremer N, Wang Y, Ishima R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (HIV-1 reverse transcriptase C38V/C280S, L289K truncated to 1 - 556) monomer, 65 kDa Human immunodeficiency virus … protein
|
| Buffer: |
50 mM Bis-Tris pH 6.8, 100 mM NaCl, 1% glycerol, 0.02% w/v NaN3, pH: 6.8
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jun 20
|
Relative domain orientation of the L289K HIV
‐1 reverse transcriptase monomer
Protein Science 31(5) (2022)
Xi Z, Ilina T, Guerrero M, Fan L, Sluis‐Cremer N, Wang Y, Ishima R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (HIV-1 reverse transcriptase C38V/C280S, L289K truncated to 1 - 556) monomer, 65 kDa Human immunodeficiency virus … protein
|
| Buffer: |
50 mM Bis-Tris pH 6.8, 100 mM NaCl, 1% glycerol, 0.02% w/v NaN3, pH: 6.8
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jun 20
|
Relative domain orientation of the L289K HIV
‐1 reverse transcriptase monomer
Protein Science 31(5) (2022)
Xi Z, Ilina T, Guerrero M, Fan L, Sluis‐Cremer N, Wang Y, Ishima R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
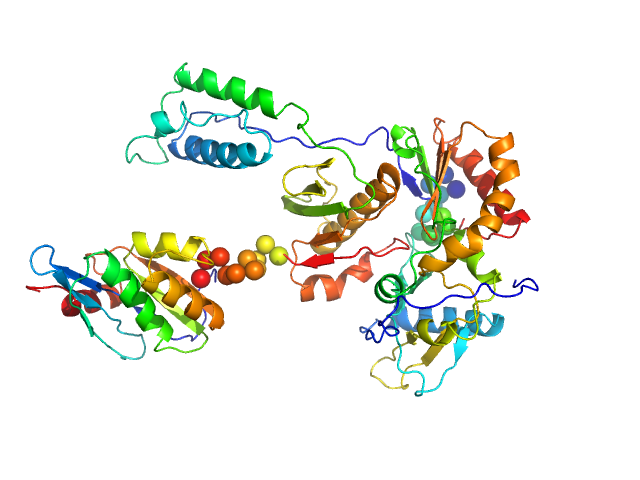
| Sample: |
Gag-Pol polyprotein (HIV-1 reverse transcriptase C38V/C280S, L289K truncated to 1 - 556) monomer, 65 kDa Human immunodeficiency virus … protein
|
| Buffer: |
50 mM Bis-Tris pH 6.8, 100 mM NaCl, 1% glycerol, 0.02% w/v NaN3, pH: 6.8
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jun 20
|
Relative domain orientation of the L289K HIV
‐1 reverse transcriptase monomer
Protein Science 31(5) (2022)
Xi Z, Ilina T, Guerrero M, Fan L, Sluis‐Cremer N, Wang Y, Ishima R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Putative autotransporter dimer, 119 kDa Escherichia coli (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 May 1
|
Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms
npj Biofilms and Microbiomes 8(1) (2022)
Vo J, Ortiz G, Totsika M, Lo A, Hancock S, Whitten A, Hor L, Peters K, Ageorges V, Caccia N, Desvaux M, Schembri M, Paxman J, Heras B
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 3
|
Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms
npj Biofilms and Microbiomes 8(1) (2022)
Vo J, Ortiz G, Totsika M, Lo A, Hancock S, Whitten A, Hor L, Peters K, Ageorges V, Caccia N, Desvaux M, Schembri M, Paxman J, Heras B
|
| RgGuinier |
4.6 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-amylase tetramer, 231 kDa Zea mays protein
|
| Buffer: |
50 mM HEPES, 25 mM NaCl, and 0.2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 7
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
615 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-amylase tetramer, 231 kDa Zea mays protein
|
| Buffer: |
50 mM HEPES, 25 mM NaCl, and 0.2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 7
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
5.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
467 |
nm3 |
|
|
|
|
|
|
|
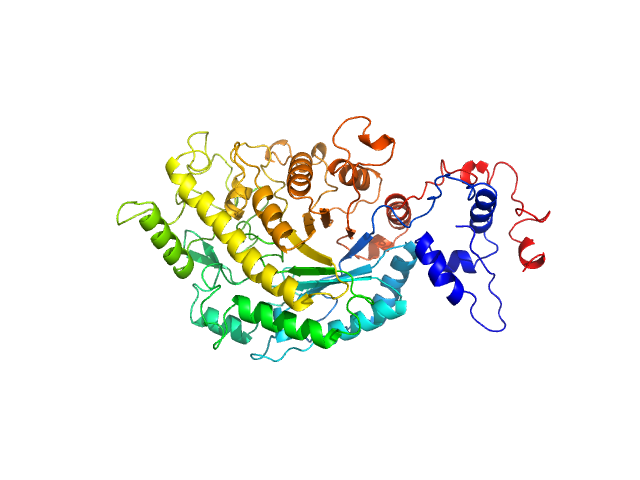
| Sample: |
Beta-amylase 1, chloroplastic monomer, 65 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM MES, 100 mM NaCl, 1 mM DTT, pH: 6.7
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 22
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
88 |
nm3 |
|
|