|
|
|
|
|
| Sample: |
Potassium voltage-gated channel subfamily B member 1 pentamer, 73 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% v/v glycerol, 0.01% Triton X-100, 10 mM β-mercaptoethanol, pH: 8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Jul 13
|
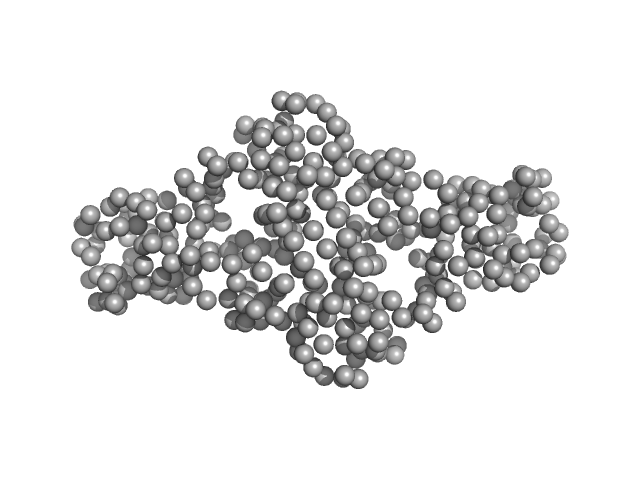
Pentameric assembly of the Kv2.1 tetramerization domain
Acta Crystallographica Section D Structural Biology 78(6):792-802 (2022)
Xu Z, Khan S, Schnicker N, Baker S
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Extracellular superoxide dismutase [Cu-Zn] dimer, 34 kDa Onchocerca volvulus protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 13
|
Crystal structure of an extracellular superoxide dismutase from Onchocerca volvulus
and implications for parasite-specific drug development
Acta Crystallographica Section F Structural Biology Communications 78(6) (2022)
Moustafa A, Perbandt M, Liebau E, Betzel C, Falke S
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
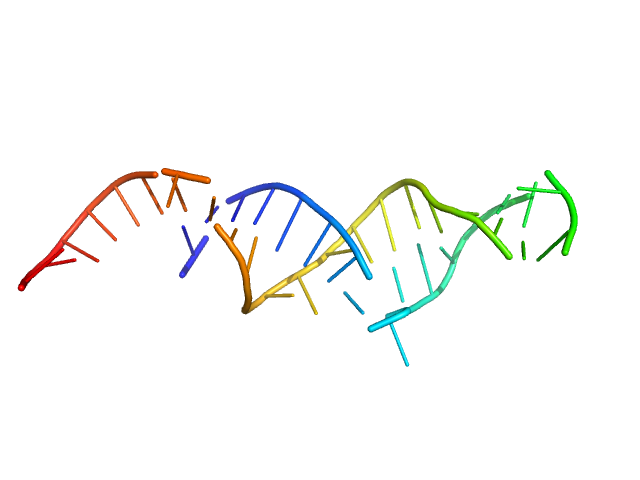
| Sample: |
Spiegelmer NOX-E36 monomer, 13 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
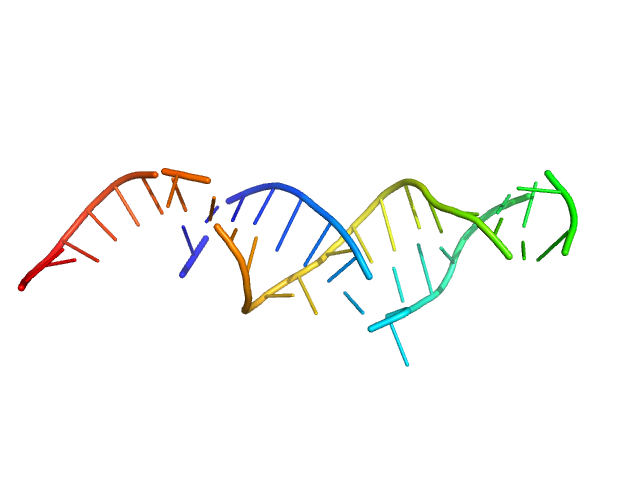
| Sample: |
Spiegelmer NOX-E36 monomer, 13 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
|
|
|
|
|
|
|
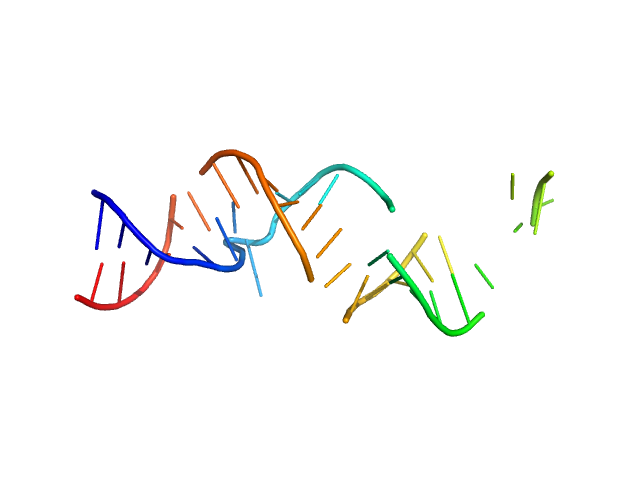
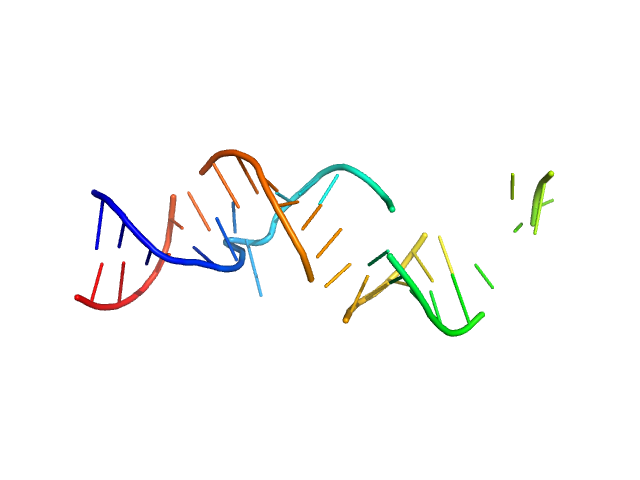
| Sample: |
Spiegelmer NOX-A12 monomer, 15 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Spiegelmer NOX-A12 monomer, 15 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
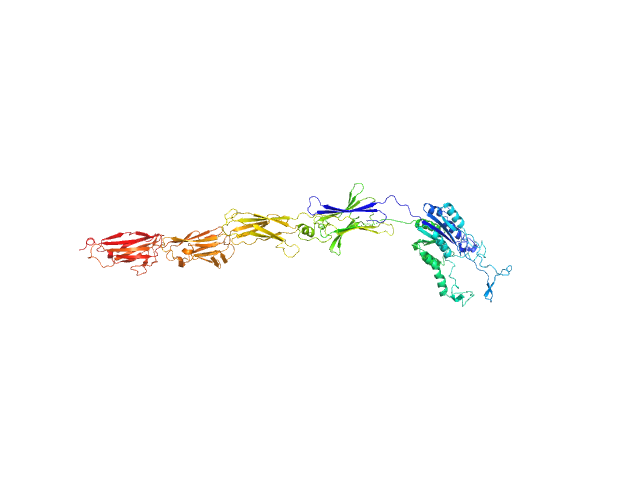
| Sample: |
von Willebrand factor type A domain protein monomer, 89 kDa Streptococcus oralis ATCC … protein
|
| Buffer: |
20 mM HEPES,150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 12
|
New structural insights into the PI-2 pilus from Streptococcus oralis, an early dental plaque colonizer.
FEBS J (2022)
Yadav RK, Krishnan V
|
| RgGuinier |
6.4 |
nm |
| Dmax |
29.2 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nocturnin monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 10% glycerol, 5 mM MgCl2, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Feb 7
|
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.
Biochemistry (2022)
Wickramaratne AC, Li L, Hopkins JB, Joachimiak LA, Green CB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nocturnin monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 10% glycerol, 5 mM MgCl2, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Feb 7
|
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.
Biochemistry (2022)
Wickramaratne AC, Li L, Hopkins JB, Joachimiak LA, Green CB
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nocturnin - Deletion construct - Δ107-120 monomer, 40 kDa protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 10% glycerol, 5 mM MgCl2, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Feb 7
|
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.
Biochemistry (2022)
Wickramaratne AC, Li L, Hopkins JB, Joachimiak LA, Green CB
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
72 |
nm3 |
|
|