|
|
|
|
|
| Sample: |
Protein jagged-1 cysteine-rich domain , 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 11
|
Notch-Jagged signaling complex defined by an interaction mosaic.
Proc Natl Acad Sci U S A 118(30) (2021)
Zeronian MR, Klykov O, Portell I de Montserrat J, Konijnenberg MJ, Gaur A, Scheltema RA, Janssen BJC
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein jagged-1 cysteine-rich domain , 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 11
|
Notch-Jagged signaling complex defined by an interaction mosaic.
Proc Natl Acad Sci U S A 118(30) (2021)
Zeronian MR, Klykov O, Portell I de Montserrat J, Konijnenberg MJ, Gaur A, Scheltema RA, Janssen BJC
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein jagged-1 cysteine-rich domain , 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 11
|
Notch-Jagged signaling complex defined by an interaction mosaic.
Proc Natl Acad Sci U S A 118(30) (2021)
Zeronian MR, Klykov O, Portell I de Montserrat J, Konijnenberg MJ, Gaur A, Scheltema RA, Janssen BJC
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurogenic locus notch homolog protein 1 full ectodomain monomer, 183 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 25
|
Notch-Jagged signaling complex defined by an interaction mosaic.
Proc Natl Acad Sci U S A 118(30) (2021)
Zeronian MR, Klykov O, Portell I de Montserrat J, Konijnenberg MJ, Gaur A, Scheltema RA, Janssen BJC
|
| RgGuinier |
10.5 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
1000 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
BON domain protein decamer, 229 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 11
|
BonA from Acinetobacter baumannii Forms a Divisome-Localized Decamer That Supports Outer Envelope Function.
mBio :e0148021 (2021)
Grinter R, Morris FC, Dunstan RA, Leung PM, Kropp A, Belousoff M, Gunasinghe SD, Scott NE, Beckham S, Peleg AY, Greening C, Li J, Heinz E, Lithgow T
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
546 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
BON domain protein monomer, 20 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 11
|
BonA from Acinetobacter baumannii Forms a Divisome-Localized Decamer That Supports Outer Envelope Function.
mBio :e0148021 (2021)
Grinter R, Morris FC, Dunstan RA, Leung PM, Kropp A, Belousoff M, Gunasinghe SD, Scott NE, Beckham S, Peleg AY, Greening C, Li J, Heinz E, Lithgow T
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
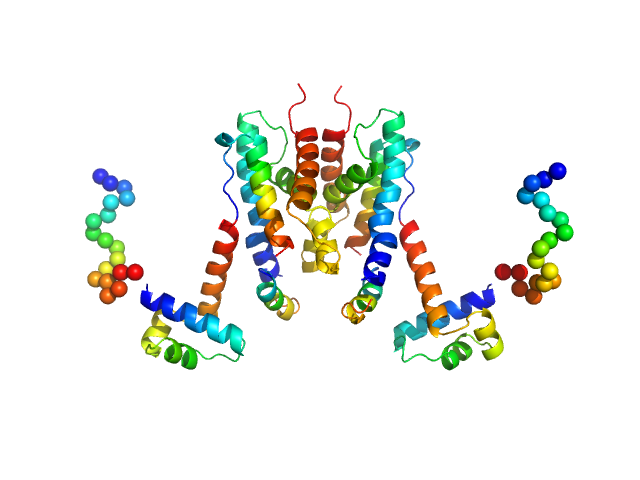
| Sample: |
TetR/AcrR family transcriptional regulator dimer, 48 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
20 mM Na2HPO4, 50 mM NaCl, 50 mM imidazole, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
The Induction Mechanism of the Flavonoid-Responsive Regulator FrrA.
FEBS J (2021)
Werner N, Werten S, Hoppen J, Palm GJ, Göttfert M, Hinrichs W
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
TetR/AcrR family transcriptional regulator dimer, 48 kDa Bradyrhizobium diazoefficiens protein
5,7-dihydroxy-3-(4-hydroxyphenyl)chromen-4-one monomer, 0 kDa
|
| Buffer: |
20 mM Na2HPO4, 50 mM NaCl, 50 mM imidazole, 0.08 mM genistein, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 27
|
The Induction Mechanism of the Flavonoid-Responsive Regulator FrrA.
FEBS J (2021)
Werner N, Werten S, Hoppen J, Palm GJ, Göttfert M, Hinrichs W
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
TetR/AcrR family transcriptional regulator dimer, 48 kDa Bradyrhizobium diazoefficiens protein
5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one monomer, 0 kDa
|
| Buffer: |
20 mM Na2HPO4, 50 mM NaCl, 50 mM imidazole, 1 mM naringenin, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
The Induction Mechanism of the Flavonoid-Responsive Regulator FrrA.
FEBS J (2021)
Werner N, Werten S, Hoppen J, Palm GJ, Göttfert M, Hinrichs W
|
| RgGuinier |
3.2 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
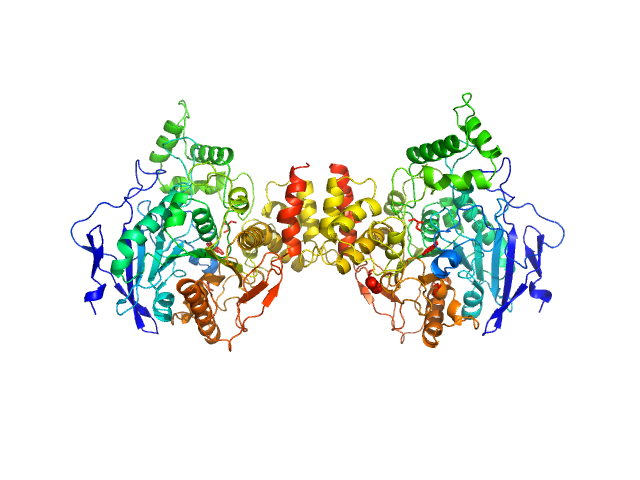
| Sample: |
acetylcholinesterase dimer, 120 kDa Homo sapiens protein
acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|
|