|
|
|
|
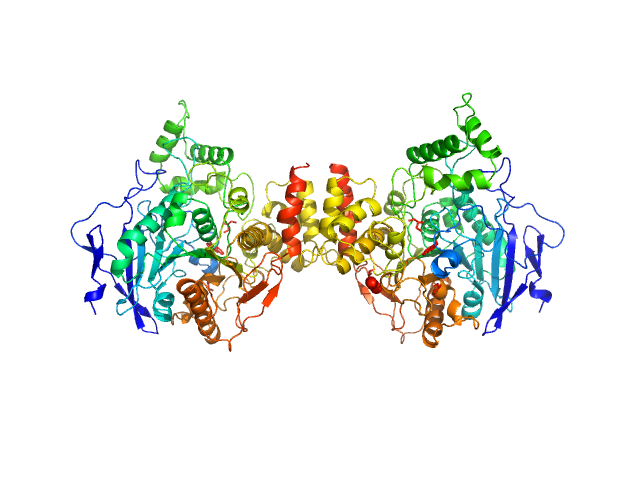
|
| Sample: |
acetylcholinesterase dimer, 120 kDa Homo sapiens protein
acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AT07459p monomer, 30 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 5 mM β-mercaptoethanol, 200 mM NaCl, 1% v/v glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Mar 8
|
Molecular insights on CALX-CBD12 inter-domain dynamics from MD simulations, RDCs and SAXS.
Biophys J (2021)
de Souza Degenhardt MF, Vitale PAM, Abiko LA, Zacharias M, Sattler M, Oliveira CLP, Salinas RK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AT07459p monomer, 30 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 5 mM β-mercaptoethanol, 200 mM NaCl, 1% v/v glycerol, 0.8 mM CaCl2, pH: 7.4
|
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Apr 13
|
Molecular insights on CALX-CBD12 inter-domain dynamics from MD simulations, RDCs and SAXS.
Biophys J (2021)
de Souza Degenhardt MF, Vitale PAM, Abiko LA, Zacharias M, Sattler M, Oliveira CLP, Salinas RK
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
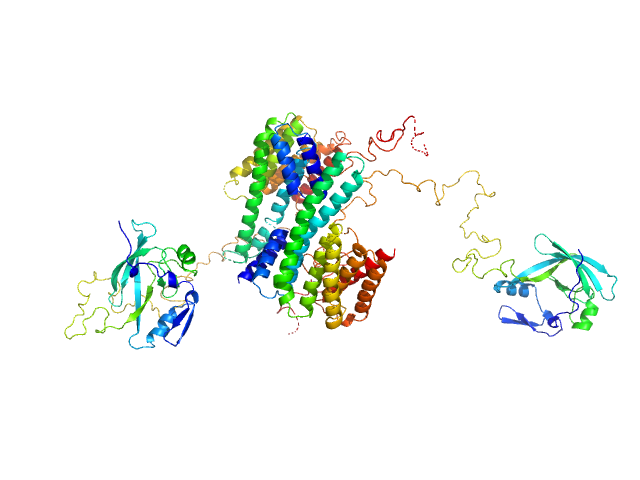
|
| Sample: |
14-3-3 protein zeta/delta dimer, 53 kDa Homo sapiens protein
Ataxin-1 AXH-C dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 13
|
A structural study of the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1.
J Mol Biol :167174 (2021)
Leysen S, Jane Burnley R, Rodriguez E, Milroy LG, Soini L, Adamski CJ, Nitschke L, Davis R, Obsil T, Brunsveld L, Crabbe T, Yahya Zoghbi H, Ottmann C, Martin Davis J
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
14-3-3 protein zeta/delta dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 13
|
A structural study of the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1.
J Mol Biol :167174 (2021)
Leysen S, Jane Burnley R, Rodriguez E, Milroy LG, Soini L, Adamski CJ, Nitschke L, Davis R, Obsil T, Brunsveld L, Crabbe T, Yahya Zoghbi H, Ottmann C, Martin Davis J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
7.9 |
nm |
|
|
|
|
|
|
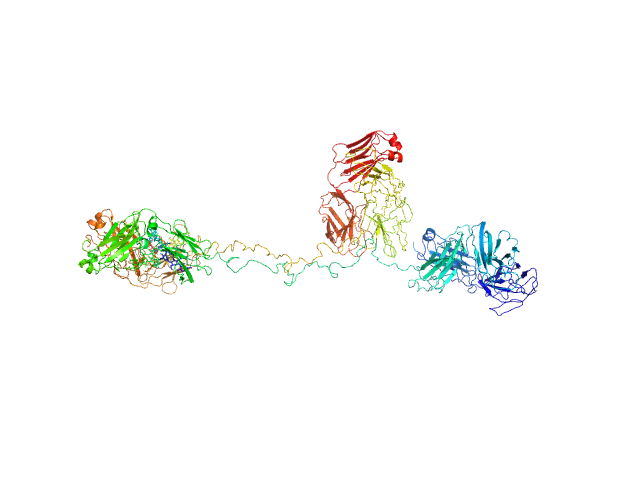
|
| Sample: |
Immunoglobulin G subclass 3 monomer, 155 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
Solution structures of human myeloma IgG3 antibody reveal extended Fab and Fc regions relative to the other IgG subclasses.
J Biol Chem :100995 (2021)
Spiteri VA, Goodall M, Doutch J, Rambo RP, Gor J, Perkins SJ
|
| RgGuinier |
7.0 |
nm |
| Dmax |
22.9 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
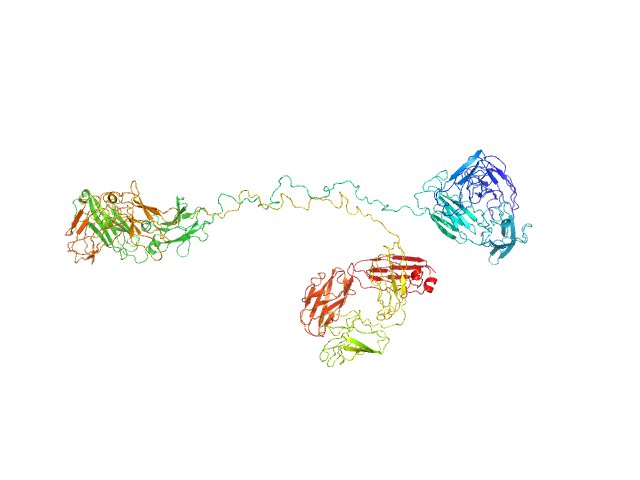
|
| Sample: |
Immunoglobulin G subclass 3 monomer, 155 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
Solution structures of human myeloma IgG3 antibody reveal extended Fab and Fc regions relative to the other IgG subclasses.
J Biol Chem :100995 (2021)
Spiteri VA, Goodall M, Doutch J, Rambo RP, Gor J, Perkins SJ
|
| RgGuinier |
7.1 |
nm |
| Dmax |
27.7 |
nm |
| VolumePorod |
294 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ataxin-1 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 13
|
A structural study of the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1.
J Mol Biol :167174 (2021)
Leysen S, Jane Burnley R, Rodriguez E, Milroy LG, Soini L, Adamski CJ, Nitschke L, Davis R, Obsil T, Brunsveld L, Crabbe T, Yahya Zoghbi H, Ottmann C, Martin Davis J
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Survival motor neuron-like protein 1 dimer, 26 kDa Schizosaccharomyces pombe (strain … protein
Survival of motor neuron protein-interacting protein yip11 dimer, 54 kDa Schizosaccharomyces pombe (strain … protein
Gcn4p dimer, 8 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 150 mM NaCl, 1 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at X21, National Synchrotron Light Source (NSLS) on 2011 Oct 21
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
6.6 |
nm |
| Dmax |
23.3 |
nm |
| VolumePorod |
320 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Survival motor neuron-like protein 1 dimer, 26 kDa Schizosaccharomyces pombe (strain … protein
Survival of motor neuron protein-interacting protein yip11 dimer, 54 kDa Schizosaccharomyces pombe (strain … protein
Gcn4p dimer, 8 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 150 mM NaCl, 1 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at X21, National Synchrotron Light Source (NSLS) on 2011 Oct 21
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
6.6 |
nm |
| Dmax |
22.9 |
nm |
| VolumePorod |
325 |
nm3 |
|
|