|
|
|
|
|
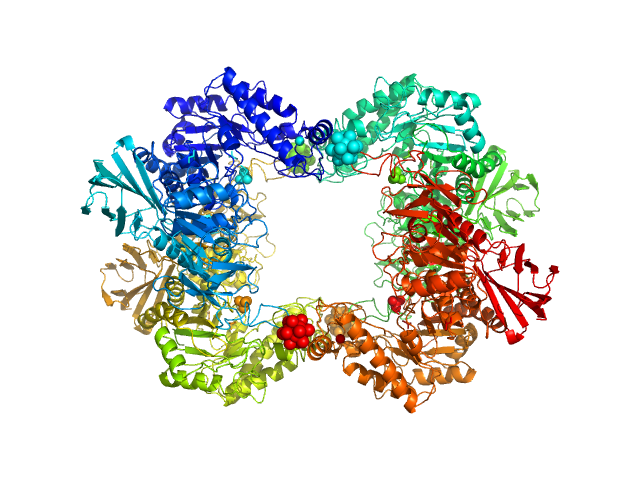
| Sample: |
Alpha-L-fucosidase H503A tetramer, 297 kDa Paenibacillus thiaminolyticus (Bacillus … protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7.4
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2019 Jul 16
|
The first structure–function study of GH151 α‐ l
‐fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
The FEBS Journal (2022)
Koval'ová T, Kovaľ T, Stránský J, Kolenko P, Dušková J, Švecová L, Vodičková P, Spiwok V, Benešová E, Lipovová P, Dohnálek J
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
433 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
11.2 |
nm |
| Dmax |
37.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
8.6 |
nm |
| Dmax |
34.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
7.2 |
nm |
| Dmax |
18.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
CUB domain-containing protein 1 monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 137 mM NaCl, 2.7 mM KCl,, pH: 7.4
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 29
|
Targeting a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) for RAS-driven cancers.
J Clin Invest 132(4) (2022)
Lim SA, Zhou J, Martinko AJ, Wang YH, Filippova EV, Steri V, Wang D, Remesh SG, Liu J, Hann B, Kossiakoff AA, Evans MJ, Leung KK, Wells JA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
185 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cleaved - CUB domain-containing protein 1 (N-terminus) monomer, 38 kDa Homo sapiens protein
Cleaved - CUB domain-containing protein 1 (C-terminus) monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 137 mM NaCl, 2.7 mM KCl,, pH: 7.4
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 29
|
Targeting a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) for RAS-driven cancers.
J Clin Invest 132(4) (2022)
Lim SA, Zhou J, Martinko AJ, Wang YH, Filippova EV, Steri V, Wang D, Remesh SG, Liu J, Hann B, Kossiakoff AA, Evans MJ, Leung KK, Wells JA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
182 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
200 mM NaCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5
|
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
RNA triplex structures revealed by WAXS-driven MD simulations
(2022)
Chen Y, He W, Kirmizialtin S, Pollack L
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
200 mM KCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5
|
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
RNA triplex structures revealed by WAXS-driven MD simulations
(2022)
Chen Y, He W, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
5 mM MgCl2, 20 mM Na-MES, 50 μM EDTA, pH: 5.5
|
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
RNA triplex structures revealed by WAXS-driven MD simulations
(2022)
Chen Y, He W, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Triplex monomer, 17 kDa RNA
|
| Buffer: |
200 mM NaCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5
|
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
RNA triplex structures revealed by WAXS-driven MD simulations
(2022)
Chen Y, He W, Kirmizialtin S, Pollack L
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
|
|